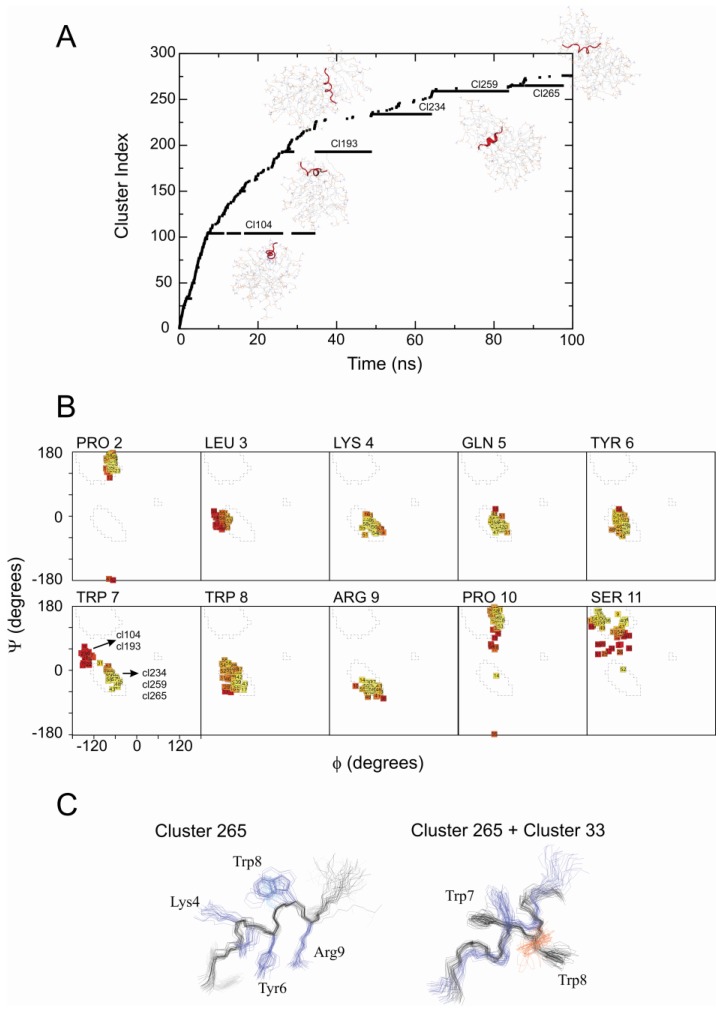
Figure 4.
Unrestrained MD simulation of PW2 interacting with free-DPC during spontaneous micelle formation. (A) Cluster index as a function of MD simulation time. From 0 to 7 ns we observed an increasing number of clusters. After 7 ns, five long-lived clusters were detected (104, 193, 234, 259 and 265) and representative snapshots of the interaction are shown for each of them. (B) Ramachandran plot of all five stable clusters for each amino acid residue. Note that the shift of the Ramanchadran position of Trp 7 from generously allowed region to helix position, indicated by arrows. (C) Superposition of 20 representative structures of cluster 265 (left) and ribbon representation of cluster 265 structure (right).