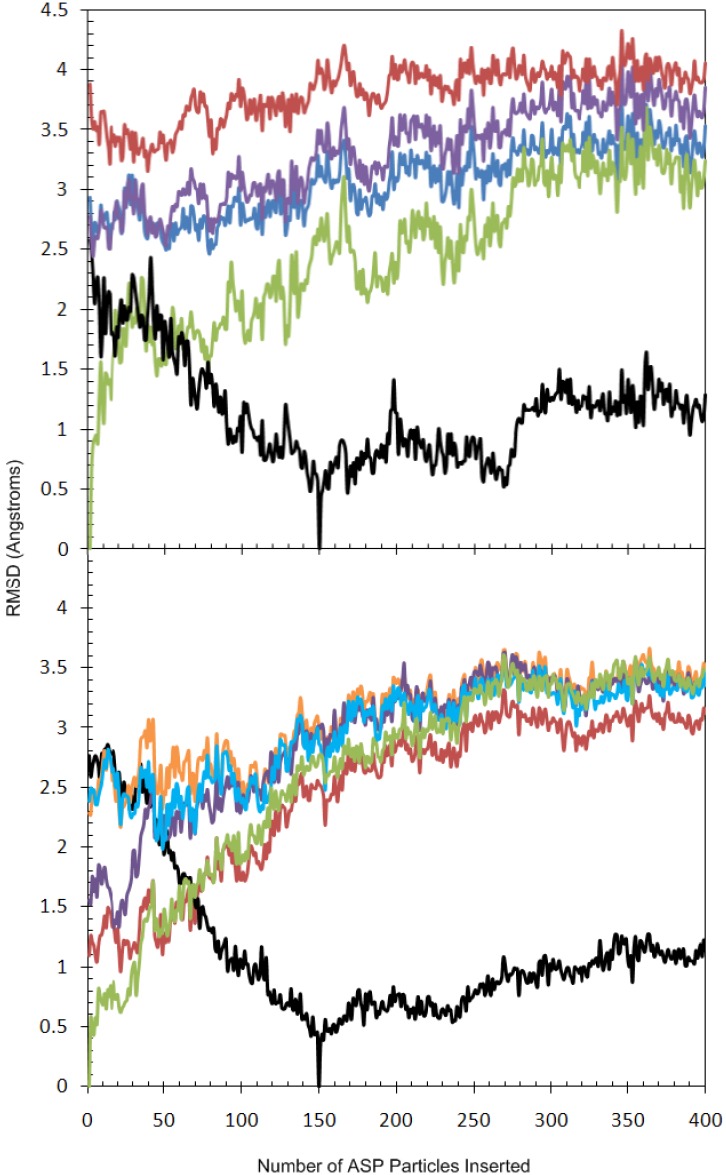
Figure 7.
(a) Evolution of the RMSD of the backbone atoms of the Gly-rich P-loop (residues 37–47) of GSK3β during the ASP simulation for 1.1 Å-spaced ASP particles, with respect to reference structures. Reference structures included the protein conformations containing 1 ASP particle (green), those containing 150 ASP particles (black), and three X-ray crystal structures for GSK3β: 1Q3W (blue), 1Q4L (purple), and 1R0E (red); (b) Evolution of the RMSD of the backbone atoms of key residues defining the CDK5 binding pocket including the residues 11–17 (the Gly-rich P-loop), 31, 33, 80–85 (the Hinge region), 130, 133 and 144–146 (the DFG motif) of CDK5 during the ASP simulation for 1.1 Å-spaced ASP particles with respect to reference structures. Reference structures included the protein conformations containing 1 ASP particle (green), those containing 150 ASP particles (black), and four ligand complexes taken from the MD simulations: R-roscovitine (red), aloisine-A (orange), indirubin-3'-oxime (purple) and ATP/Mg2+ (cyan).