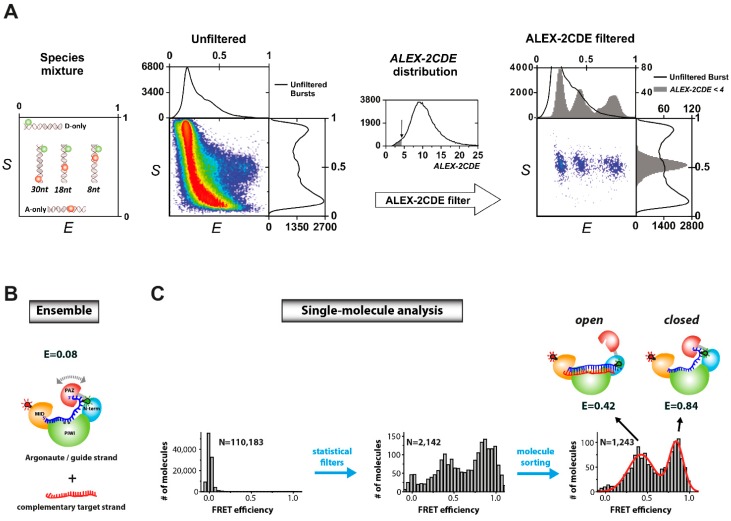
Figure 8.
(A) Advanced statistical analysis of single-molecule FRET data acquired using confocal microscopy with alternating laser excitation (ALEX) [142]. The ALEX-2CDE filter uncovers a minor, doubly labeled dsDNA subpopulation hidden in the larger concentrations of singly labeled dsDNA subpopulations [30 pM donor-labeled and 30 pM acceptor-labeled ssDNA with three different doubly labeled dsDNA, 6 pM each (E = 0.22, E = 0.46, and E = 0.85)]. (Adapted from Tomov, T. E. et al., 2012 “Disentangling subpopulations in single-molecule FRET and ALEX experiments with photon distribution analysis”. Biophysical Journal 102(5): 1163–1173); (B) In order to understand the conformational transitions of the Argonaute-protein a donor-acceptor pair (AttoATTO550/Alexa647) was engineered into the protein-nucleic acid complex. In ensemble measurements a surprisingly low FRET efficiency of 0.08 was determined; (C) When measured at the single-molecule level a comparable low FRET efficiency was measured. However, unlike in ensemble, the single-molecule approach revealed that the low FRET efficiency is due to a large access of donor-only and acceptor-only labeled molecules in the sample. Statistical filters gave access to the 1% of molecules that exhibit a stable FRET signal. Ultimately, this revealed that Argonaute adopts a different conformation (E = 0.42) when the target strand is loaded as compared to the Argonaute-guide DNA only complex, which is characterized by a high FRET value (E = 0.84).