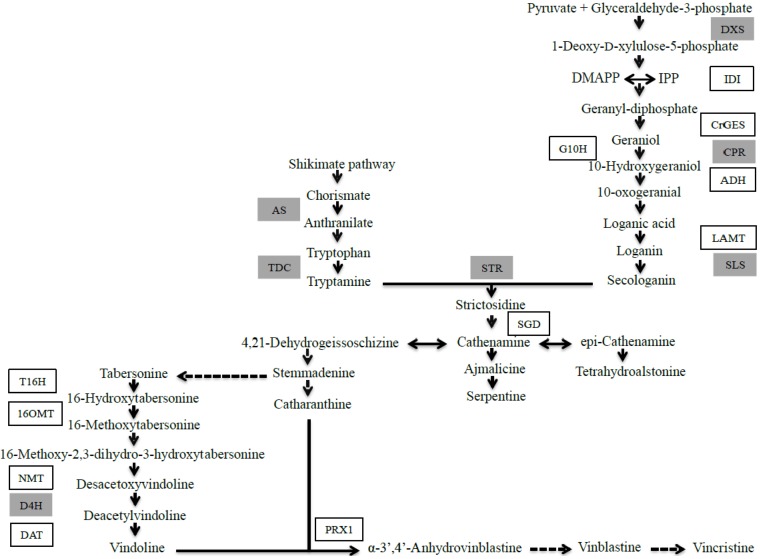
Scheme 1.
Biosynthetic pathway of TIA in C. roseus (modified from [20]). Broken arrows indicate multiple enzymatic reactions. Enzymes with their corresponding cloned gene are indicated. Genes previously reported to be regulated by ORCA3 are indicated in gray boxes.
ADH: acyclic monoterpene primary alcohol dehydrogenase; AS: anthranilate synthase; CPR: cytochrome P450 reductase; D4H: desacetoxyvindoline 4-hydrolase; DAT: acetyl-CoA:4-O-deacetylvindoline 4-O-acetyltransferase; DXS: 1-deoxy-d-xylulose-5-phosphate synthase; G10H: geraniol 10-hydroxylase; CrGES: geraniol synthase; IDI: isopentenyl diphosphate isomerase; LAMT: S-adenosyl-l-methionine:loganic acid methyltransferase; NMT: 16-methoxy-2,3-dihydro-3-hydroxytabersonine; 16OMT: 16-hydroxytabersonine-16-O-methyltransferase; PRX1: peroxidase 1; SGD: strictosidine β-d-glucosidase; SLS: secologanin synthase; STR: strictosidine synthase; TDC: tryptophan decarboxylase; T16H: tabersonine 16-hydroxylase.