Abstract
The Cre/loxP site-specific recombination system was applied to Aurantiochytrium limacinum to obtain a transformant without the antibiotic resistance marker gene. First, the enhanced green fluorescent protein gene (egfp) and chloramphenicol resistance gene (Cmr), along with the two loxP loci, were integrated into the genome of A. limacinum OUC88 using 18S rDNA sequences as the homologous recombination sites. Then plasmid pSH65, containing a zeocin resistance gene (Bler) was transferred into A. limacinum OUC_CG. After induction with galactose, repeated passage in culture and PCR-based assessment, the pSH65 plasmid was lost and A. limacinum OUC_EG host was shown to no longer have resistance to 100 mg chloramphenicol/L or 5 mg zeocin/L. Through southern blotting and fluorescence detection, egfp was found to be integrated into the genome of A. limacinum OUC_EG, and EGFP was successfully expressed in the cells. The successful application of the Cre/loxP system demonstrates an experimental basis for genetic modification of A. limacinum so as to obtain transformed strains with no antibiotic resistance marker genes.
Keywords: antibiotic resistance marker gene, Aurantiochytrium limacinum, Cre/loxP site-specific recombination system, homologous recombination
1. Introduction
In recent years, a genetic transformation method has been successfully established in Aurantiochytrium limacinum. A zeocin resistance gene was introduced into A. limacinum using particle bombardment [1]. Cheng et al. integrated the zeocin resistance gene into the 18S rDNA sequences of A. limacinum using electrotransformation and homologous recombination technology [2]. Agrobacterium tumefaciens-mediated transformation was also successfully applied to A. limacinum and the results demonstrated that the exogenous egfp gene had been successfully incorporated into the genome and that it was expressed efficiently [3]. Furthermore, a partial sequence of the pks gene in A. limacinum was successfully knocked out by electroporation, using the 18S rDNA sequences of A. limacinum as homologous recombination sites [4]. However, all these A. limacinum transformants retained antibiotic resistance genes, which is adverse for the industrial application of A. limacinum and was environmentally undesirable. Thus, research on a new method to eliminate the antibiotic resistance genes in genetic transformations of A. limacinum is of significant importance.
The Cre/loxP site-specific recombination system was first developed in phage p1 of Escherichia coli [5,6] and consists of the Cre recombinase and two loxP loci. In the Cre/loxP site-specific recombination system, there are two important steps: first, the circular plasmid containing two loxP loci are transferred into a host, and then the circular plasmid pSH65 with the Cre recombinase is transferred into the host, where it specifically recognizes the loxP sites [7]. Depending on the directivity of the loxP site, specific inversion, elimination or translocation of the target fragment is performed by the Cre recombinase. The advantage of the Cre/loxP site-specific recombination system is that it can specifically eliminate the antibiotic resistance gene markers that are transformed into the host in the first step, and the pSH65 plasmid can then be lost by repeated sub-culturing of the host after the second step. In 1990, the Cre/loxP recombination system was applied successfully in tobacco for site-specific reorganization, which was the first application of the Cre/loxP recombination system in plant cells [8]. Subsequently, the Cre/loxP recombination system was used to remove the marker genes from genetically modified organisms. This has been performed successfully in yeast and other organisms [9,10,11,12,13]. In this paper, the Cre/loxP site-specific recombination system was, for the first time, applied to genetic transformation of A. limacinum, where it has provided the experimental basis for directional transformation of A. limacinum without leaving behind an antibiotic marker.
2. Results and Discussion
2.1. Construction of plasmid p18SCPGC
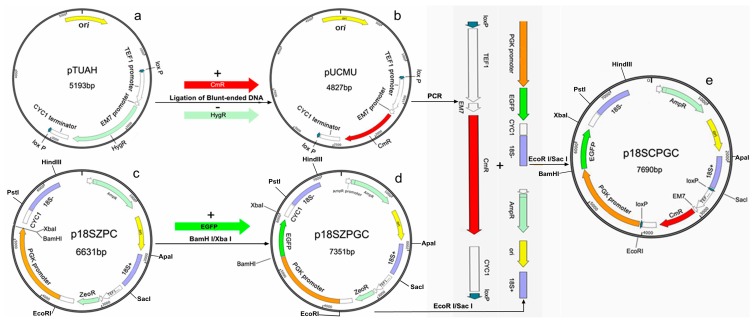
Plasmid p18SCPGC was successfully constructed using the procedure described in the Experimental section. “0” point was a reference point relative to the location of all genes and sequences (Figure 1e). After sequencing and alignment, it was established that the two homologous 18S rDNA sequences of A. limacinum were located at 1893–2507 (615 bp of 18S+) and 6605–7284 (680 bp of 18S−), respectively, which constituted the two ends of the transformation fragments. The two loxP sequences were located at 2514–2547 and 4017–4050 respectively, and flank the Cmr antibiotic resistance gene. The coding sequence of the Cmr gene was at 3032–3691 with the TEF1 promoter and the CYC1 terminator at the two ends. The PGK promoter sequence was at 4057–5548, which was followed by the coding sequence of the egfp gene at 5555–6275. All the sequences mentioned above are consistent with the expectations, which demonstrated that plasmid p18SCPGC was constructed correctly.
Figure 1.
Construction process of the plasmid. (a) Plasmid pYUAH; (b) Plasmid pUCMU; (c) Plasmid p18SZPC; (d) Plasmid p18SZPGC; (e) Plasmid p18SCPGC.
2.2. Screening of the Transformant after the First Electrotransformation Step
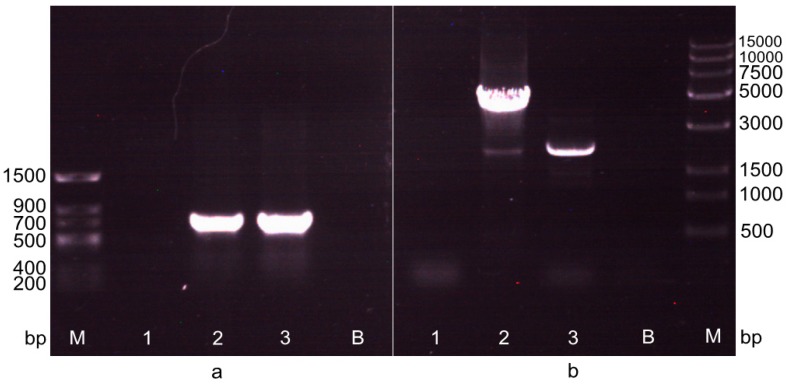
Plasmid p18SCPGC was transformed into A. limacinum OUC88, and a transformant, named A. limacinum OUC_CG, was selected on solid medium containing chloramphenicol. The Cmr antibiotic marker gene, integrated into the genome of A. limacinum OUC_CG, was detected by PCR using the Cm-F/R primer, and the genomic DNA of A. limacinum OUC88, OUC_CG and pACYCDuet-1 plasmid as the templates, respectively. Electrophoresis results (Figure 2a) showed that the amplified fragment had a size of between 500 bp to 750 bp, which is consistent with the length of the Cmr gene (660 bp). After sequencing, the amplified fragment was exactly the Cm resistance gene, which showed that A. limacinum OUC_CG was a positive transformant.
Figure 2.
The detection of recombinant in the two transformations by PCR amplification. (a) The detection of recombinant in the first transformation by PCR amplification with primers Cm-F/R. M: Trans DNA Marker II. 1: Negative control, the PCR amplification band of Cmr gene with the genomic DNA of A. limacinum OUC88 as the template. 2: The PCR amplification band of Cmr gene with the genomic DNA of A. limacinum OUC_CG as the template. 3: Positive control, the PCR amplification band of Cmr gene with the plasmid pACYCDuet-1 as the template. B: Blank control; (b) The detection of recombinant in the second transformation by PCR amplification with primers 18S+-F/Ppgk-R. M: Trans15K DNA Marker. 1: The PCR amplification band with genomic DNA of A. limacinum OUC88 as the template; 2: The PCR amplification band (3655 bp) of 18S+-loxP-Cmr-loxP-Ppgk with genomic DNA of A. limacinum OUC_CG as the template; 3: The PCR amplification band (2185 bp) of 18S+-loxP-loxP-Ppgk with genomic DNA of A. limacinum OUC_EG as the template; B: Blank control.
2.3. Screening of the Transformant in the Second Electrotransformation Step
Plasmid pSH65 was transformed into cells of A. limacinum OUC_CG by electroporation, and transformants were selected on solid medium containing both zeocin and chloramphenicol. Then the transformants were cultured in a liquid galactose induction medium at 23 °C for 48 h and plated onto solid medium containing zeocin. After that, the induced transformants were inoculated onto two solid media containing zeocin and chloramphenicol, respectively. The positive transformants grew normally on the zeocin solid medium, but were unable to grow on the chloramphenicol solid medium, indicating that the Cmr gene has been deleted from recombinants in the initial step.
The recombinants were detected by PCR and sequencing using the 18S+-F/Ppgk-R primer (Table 1). The result showed that the Cmr gene, located between the two loxP sites, had been successfully removed from A. limacinum OUC_CG by the action of Cre recombinase (Figure 2b), and further confirmed that the GAL1 promoter of S. cerevisiae on the pSH65 plasmid could play the role of promoting the expression of Cre in A. limacinum. The desired candidate transformant was sub-cultured in medium without antibiotics and the cultures were spread on two solid media, with and without zeocin, respectively. When the culture could no longer grow on zeocin solid medium, the final recombinant had been obtained and was named A. limacinum OUC_EG.
Table 1.
Primer pairs used to amplify genes described in this study.
| Primers | Sequence (from 5ʹ to 3ʹ) | Template | Product | Length (bp) |
|---|---|---|---|---|
| Cm-F | ATGGAGAAAAAAATCACTGGAT | Plasmid pACYCDuet-1 | Cmr | 660 |
| Cm-R | TTACGCCCCGCCCTGCCACTCA | |||
| pTUAH-F | CACGTCCGACGGCGGCCCGAC | Plasmid pTUAH | pTUAH | 4168 |
| pTUAH-R | GGTTTAGTTCCTCACCTTGTCG | |||
| EGFP-F | CGCGGATCCATGGTGAGCAAGGGCGAGGA | Plasmid pEGFP-1 | egfp | 720 |
| EGFP-R | TGCTCTAGATTACTTGTACAGCTCGTCCA | |||
| loxP-F | CGAGCTCCTGCTAACATCAAAAGGCCT | Plasmid pUCMU | loxP-Cmr-loxP | 1536 |
| loxP-R | CCGGAATTCATCTTGACTGATTTTTCCATGG | |||
| 18S+-F | GCGGGGCCCGTAGTGTACTGGACTACGGTG | Genomic DNA of different transformants | 18S+-loxP-Cmr-loxP-Ppgk | 3655 |
| Ppgk-R | CGCGGATCCATATTTGTTGTAAAAAGTAGATAATTAC | 18S+-loxP-loxP-Ppgk | 2185 |
Note: The underlined portions of the primer sequence show the restriction enzyme cutting sites.
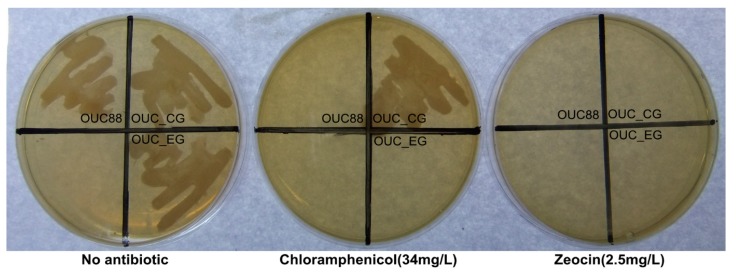
The A. limacinum OUC88, OUC_CG and OUC_EG strains were inoculated on three solid media, no antibiotics, 100 mg chloramphenicol/L and 5 mg zeocin/L respectively (Figure 3). A. limacinum OUC_CG was able to grow on solid medium without antibiotics or on medium with chloramphenicol, but could not grow on solid medium containing zeocin. There were no strains that grew on the medium containing zeocin. A. limacinum OUC88 and OUC_EG could grow normally only on medium without antibiotics. The results indicated that the antibiotic resistance genes had been successfully removed from the recombinant A. limacinum.
Figure 3.
The growth of A. limacinum OUC88, A. limacinum OUC_CG and A. limacinum OUC_EG on different solid media.
2.4. Hybridization Detection in Southern Blotting
The genomic DNA of A. limacinum OUC88, OUC_CG and OUC_EG were extracted using a Yeast DNAiso Kit. Through agarose electrophoresis, the genomic DNAs of all the samples were found to be intact and of high purity, and thus could be used in southern blotting.
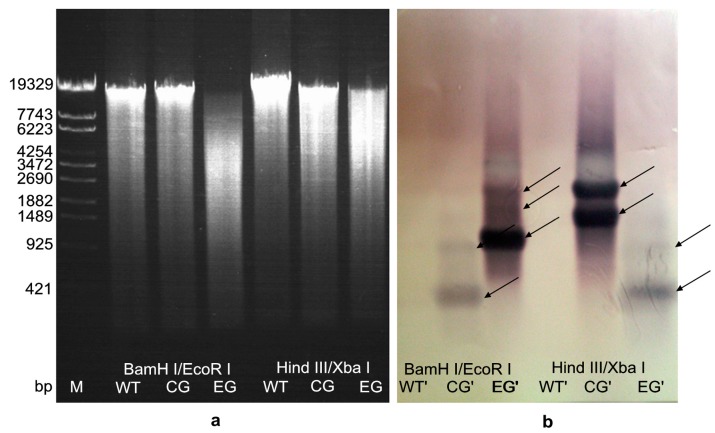
Genomic DNAs of all samples were individually digested by BamHI/EcoRI and XbaI/HindIII enzymes respectively, and the digestion fragments were homodispersed by agarose electrophoresis (Figure 4a). The hybridization signals for southern blotting were obtained using egfp as the probe (Figure 4b). A. limacinum OUC88 had no hybridization signal. For A. limacinum OUC_CG and OUC_EG, more than one hybridization bands were found after digestion with either BamHI/EcoRI or HindIII/XbaI. These bands represent the presence of egfp fragments, thus the results indicate that the egfp gene had been integrated into the genome of A. limacinum OUC_CG and OUC_EG. The southern blotting results showed that the copy number of egfp gene in the genome of A. limacinum OUC_CG and OUC_EG was at least one-copy, and further confirmed that the egfp in A. limacinum OUC_EG was still present after the Cmr gene had been removed from A. limacinum OUC_CG.
Figure 4.
Southern blotting analysis of egfp in the genomic DNA of transformants (a) The results of total DNA digested by restriction enzymes. Molecular-size of markers (bp) is shown on the left. The genomic DNA of A. limacinum OUC88 (WT), A. limacinum OUC_CG and A. limacinum OUC_EG were double digested with either BamHI/EcoRI or HindIII/XbaI; (b) The southern-blot hybridization results for the egfp gene. The number of fragment copies as deduced from the comparison of the hybridization bands are depicted for the transformants.
2.5. Fluorescence Detection of Recombinants
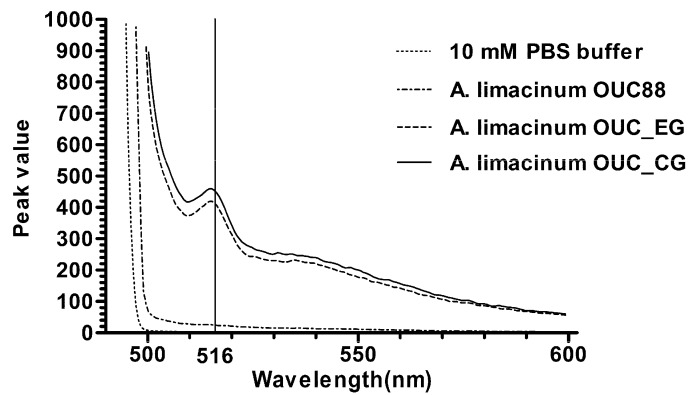
With 10 mM PBS buffer as the blank control, and A. limacinum OUC88 as the negative control, the fluorescence spectra for A. limacinum OUC_CG and OUC_EG were determined. As shown in Figure 5, fluorescence was detected in both A. limacinum OUC_CG and OUC_EG, with excitation at 488 nm and an emission peak at 516 nm, which is the characteristic fluorescence of EGFP. Thus, EGFP was expressed in A. limacinum OUC_CG and OUC_EG, which proves that the egfp gene was integrated into the genome of A. limacinum in the first electroporation and its expression was unchanged after the Cmr gene had been removed from A. limacinum OUC_CG in the second electroporation.
Figure 5.
Fluorescence measurements for the recombinants.
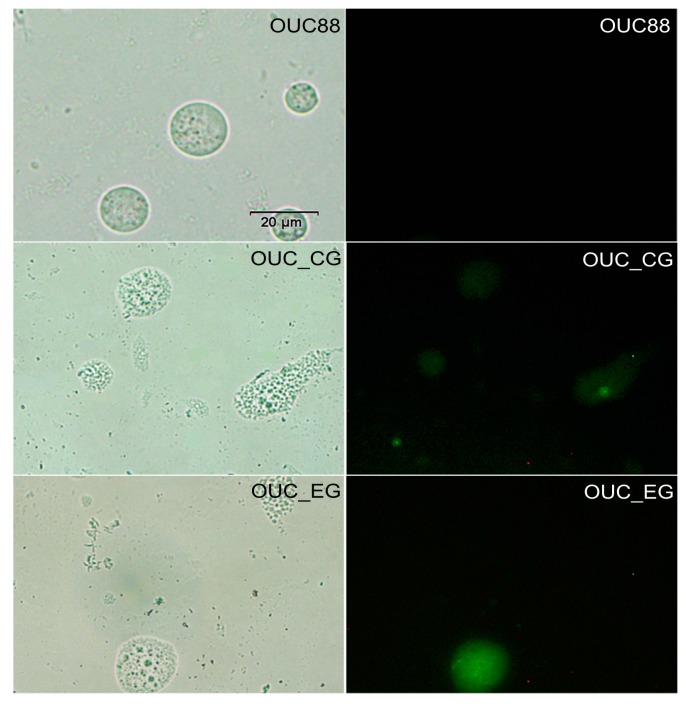
Finally, the transformants were examined by fluorescence microscopy. The results showed EGFP was expressed stably in A. limacinum OUC_CG and OUC_EG, confirming the expression of the egfp gene in the transgenic A. limacinum (Figure 6).
Figure 6.
Fluorescence microscopic analysis of the A. limacinum transformant (bars = 20 μm).
3. Experimental Section
3.1. Strains and Media
The A. limacinum OUC88 strain was preserved in the China General Microbiological Culture Collection Center (CGMCC No.1240, Beijing, China) and used as the host for the transformation experiment. Solid medium (6% (w/v) glucose, 2% (w/v) yeast extract and 2% (w/v) agar) with a salinity equivalent to 50% that of seawater was used for the conservation and selection of A. limacinum transformants at 23 °C; Liquid medium (7% (w/v) glucose, 2% (w/v) yeast extract and 2% (w/v) sodium glutamate) with a salinity equivalent to 50% that of seawater was used for the propagation of A. limacinum at 23 °C. Liquid galactose induction medium was used to induce the Cre enzyme responsible for removing Cmr from the transformants. Galactose replaced glucose in this liquid medium.
3.2. Plasmids
The plasmids pEGFP-1 and pACYCDuet-1 were purchased from Clontech (Otsu, Japan, Cat.No.6086-1) and Novagen (Billerica, MA, USA, Cat.No.71147-3) respectively; Plasmids pTUAH (Figure 1a) and pSH65 were a gift from Z.M. Chi (Ocean University of China, Qingdao, China.). The pSH65 plasmid contained the sequences of the Cre recombinase gene, GAL1 promoter and TEF1 promoter of Saccharomyces cerevisiae, CYC1 terminator, Bler and Ampr resistance genes. It was useful to express the Cre recombinase under the control of galactose and markers flanked loxP would be removed from the genome of transformant [14,15]. At the same time, the plasmid pSH65 was unstable and would be disappeared in transformants when transformants were subcultured reach over a certain number of times.
The plasmid p18SZPC (Figure 1c) was constructed in our previous work based on plasmid pTEF1/Zeo (V50320, Invitrogen, Carlsbad, CA, USA), which contained sequences of the 18S rDNA of A. limacinum (GenBank: HM042908.2), the PGK promoter of S. cerevisiae, the CYC1 terminator, and antibiotic resistance genes Bler and Ampr.
3.3. Construction of the Plasmid p18SCPGC
The plasmid p18SCPGC was constructed to contain 18S rDNA sequences, egfp as a reporter gene, the Cmr antibiotic resistance gene and two loxP sites. The 18S rDNA sequences were used as the homologous recombination sites to integrate the heterologous genes into A. limacinum. The egfp reporter gene was used to detect the expression of the heterologous gene in A. limacinum. The Cmr gene was used for screening transgenic strains of A. limacinum, and was located between the two loxP sites. The Cmr gene was subsequently removed by the inducible expression of Cre recombinase.
(1) The plasmid pTUAH without Hygr was amplified from pTUAH by PCR with primer pair pTUAH-F/R (Table 1). The primer pair Cm-F/R (Table 1) was used to amplify the antibiotic resistance gene, Cmr, from the vector pACYCDuet-1. The resistance gene Cmr and the vector pTUAH were ligated to each other and the resulting vector was designated as plasmid pUCMU (Figure 1b).
(2) The coding region of egfp was amplified from vector pEGFP-1 with primers EGFP-F/R (Table 1) and introduced into the plasmid p18SZPC between BamHI and XbaI, and the resulting vector was named plasmid p18SZPGC (Figure 1d).
(3) The fragment loxP-TEF1-EM7-Cmr-CYC1-loxP was obtained from the plasmid pUCMU by PCR with primer pair loxP-F/R (Table 1) and ligated into plasmid p18SZPGC between EcoRI and SacI. The resulting vector was named plasmid p18SCPGC (Figure 1e).
3.4. Electrotransformation
The circular plasmid was transformed into A. limacinum by electrotransformation following the procedure described by Cheng [2]. The cells of A. limacinum OUC88 were cultured overnight in 10 mL liquid medium, inoculated into 50 mL fresh medium and grown to a logarithmic phase (OD600 ≈ 1.5). Cells were harvested by centrifugation at 8000× g for 5 min at 4 °C. The resuspended pellet was washed sequentially with 10 mL ice-cold sterile water and 10 mL of 1 M sorbitol solution, and then diluted to 106 cells/mL with 1 M sorbitol solution. The A. limacinum competent cells were mixed with circular plasmid DNA and transferred to a 0.2-cm cuvette for electroporation. The most suitable electroporation parameters were: 1.8 kV/cm, 200 Ω and 50 μF. After electroporation, the solution was recovered in 1 mL liquid medium and incubated at 28 °C, 200 rpm/min, for 1 h.
3.5. The Screening of Transformants
A. limacinum was not sensitive to ampicillin, kanamycin or streptomycin, but sensitive to chloramphenicol and zeocin [16]. Even a high concentration of the former group of antibiotics (50–300 mg/L) could not inhibit the growth of A. limacinum and its survival rate was still over 80%. However, the survival rate of A. limacinum cultured with chloramphenicol was below 13% at a concentration of 25.5–68 mg/L, and only 2.10%–2.79% at a zeocin concentration of 2.5–4.0 mg/L. Accordingly, 100 mg chloramphenicol/l and 5 mg zeocin/L were used respectively for screening of transformants after the two transformation steps in this research.
In the first step, plasmid p18SCPGC was transformed into A. limacinum OUC88, which integrated the loxP-Cmr-loxP-egfp fragment into the genome by homologous recombination with the 18S rDNA sequences of A. limacinum OUC88. The recombinants were screened on solid medium containing 100 mg chloramphenicol/L, and were confirmed by PCR amplification of the Cmr gene. After sequencing of the amplified Cmr fragment, the recombinant was used in the following step.
In the second step, plasmid pSH65 was transformed into the previous recombinant. After selection on solid medium with zeocin and incubation at 23 °C for 48 h in the dark, the transformants were cultured in liquid galactose induction medium in the dark (200 rpm, 23 °C, 48 h) to induce the expression of Cre recombinase in plasmid pSH65 so as to excise the Cmr gene located between the loxP sites. Subsequently, the transformants were inoculated simultaneously on two solid media containing chloramphenicol and zeocin, respectively. One reombinant that could grow on zeocin solid medium, but could not grow on chloramphenicol solid medium, was selected as a candidate recombinant. This candidate recombinant was tested by PCR, using the primers Cm-F/R to confirm the deletion of the Cmr gene.
Finally, the candidate recombinant was sub-cultured in medium without antibiotics for 10 generations during which plasmid pSH65 could be lost [17]. The cultures were spread on two solid media with and without zeocin, respectively. When the culture could no longer grow on zeocin solid medium, the final recombinant had been obtained.
3.6. Southern Blotting of the Recombinants
Genomic DNA was extracted from A. limacinum OUC88 and the transformants respectively using a Yeast DNAiso Kit (D9082, Takara, Otsu, Japan). DNA samples were digested with two groups of restriction enzymes (BamHI/EcoRI, XbaI/HindIII) and for each sample, the resultant DNA fragments were separated on an 0.8% agarose gel and transferred to a nitrocellulose membrane (0.22 μm, Pall, New York, NY, USA) by siphon blotting. The DNA fragment for egfp, used as the probe, was prepared by PCR using the primers EGFP-F/R. Subsequently, the purified fragments were labeled with DIG before use as DNA hybridization probes. Probe detection in the Southern blotting was performed using a DIG High Prime DNA Labeling and Detection Starter Kit I (for color detection with NBT/BCIP) (Cat.NO.11745832910, Roche, Basel, Switzerland).
3.7. Fluorescence Detection of the Recombinants
The A. limacinum OUC88 and transformants were cultured overnight in the liquid medium, inoculated into fresh medium and grown to logarithmic phase. Cells were harvested by centrifugation at 8000× g for 5 min at 4 °C. The collected cells were washed twice with sterile water and re-suspended in 10 mM PBS buffer (pH 7.2). Cell suspensions of A. limacinum were diluted to 106 cells/mL with 10 mM PBS buffer and the fluorescence spectrum was measured using an F4500 fluorescence spectrophotometer (Hitachi, Tokyo, Japan). Detection parameters for measuring the fluorescence spectrum were as follows: scanning speed 1200 nm/min, slit width 5.0 nm, voltage 950 V and excitation wavelength 488 nm.
4. Conclusions
A. limacinum is an ideal industrial species for producing DHA [18,19,20,21,22], and its application prospects are very broad [23,24,25]. Optimization of culture conditions and breeding of mutants have been common techniques to improve the yield of DHA in A. limacinum; However, these methods cannot not improve the content of DHA readily or in a controlled manner. At present, genetic engineering has become an effective method to change genetically-based biological traits. The greatest advantage of genetic engineering is that it is highly specific, and use of this technology can avoid problems, such as the uncertain nature of mutations and the work-load of screening in traditional mutation breeding. In A. limacinum, the methods for genetic transformation have already been established. Electrotransformation and Agrobacterium tumefaciens-mediated transformation have been successfully applied to A. limacinum. Homologous recombination also has been shown to be effective in integrating a foreign gene into the genomic DNA of A. limacinum. In order to make the transformants suitable for industrial application, the selectable antibiotic resistance marker genes should be removed. Accordingly, the Cre/loxP system has been applied to A. limacinum for the first time in this research.
First, two loxP loci, the chloramphenicol resistance gene and the egfp gene were integrated into the genome of A. limacinum OUC88, using the 18S rDNA sequences of A. limacinum as the homologous recombination sites. Verified by antibiotic resistance screening, PCR and sequencing, a transformant named A. limacinum OUC_CG was obtained. Next, the plasmid pSH65 was incorporated into A. limacinum OUC_CG and Cre recombinase was expressed by galactose induction. After antibiotic resistance screening, A. limacinum OUC_EG was found to grow on the solid medium containing 5 mg zeocin/L, but not on the solid medium having 100 mg chloramphenicol/L. As proven by PCR, the Cmr sequence located between the two loxP loci had been successfully removed from the genome of A. limacinum OUC_EG. During continuous sub-culturing on medium without antibiotics, the recombinant A. limacinum OUC_EG lost the plasmid pSH65 and completely lost its antibiotic resistance. Subsequently, through southern blotting, egfp was proven to have integrated into the genome of A. limacinum with a copy number of at least one. By fluorescence detection of EGFP, A. limacinum OUC88 had no emission peak, whereas A. limacinum OUC_CG and OUC_EG both had an obvious emission peak at 516 nm after excitation at 488 nm. These observations suggest that EGFP was expressed in A. limacinum OUC_CG and OUC_EG, and that the presence of egfp was not affected by the deletion of the Cmr gene by the action of Cre recombinase.
In this paper, the Cre/loxP site-specific recombination system was introduced into A. limacinum OUC88. Using the 18S rDNA sequences as the homologous recombination sites, egfp was integrated into the genome of A. limacinum and expressed successfully. In addition, the antibiotic resistance gene was eliminated from A. limacinum. This research has laid an experimental foundation for using genetic engineering technology to modify the traits of A. limacinum and obtain transformed strains that have no antibiotic resistance marker, which make them potentially useful for applications in industry.
Acknowledgments
This research was supported by the Research Award Fund for Young and Middle-aged Scientists of Shandong Province (Grant No.BS2012HZ017) and the Research Fund for the National High Technology Research and Development Program of China (Grant No. 2008AA09Z410).
We are grateful to Z.M. Chi (Ocean University of China, Qingdao, China) for technical advice and kindly providing the plasmids pTUAH and pSH65. We thank G.C. Wang (Institute of Oceanology, Chinese Academy of Science, Qingdao, China) for technical advice and support concerning analysis of fluorescence spectra using a F4500 fluorescence spectrophotometer (Hitachi, Tokyo, Japan). We would also like to thank John P. van der Meer for his comments and support during the writing of this manuscript.
Author Contributions
H Sun, H Chen and P Hou conceived and designed the study. B Zhou, Y Liu, F Wu and X Cao performed the experiments. H Sun wrote the paper. X Zang and X Zhang reviewed and edited the manuscript. All authors read and approved the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
Footnotes
Sample Availability: The strain of A. limacinum OUC88 and all the primers used in this research are available from the authors.
References
- 1.Lippmeier J.C., Crawford K.S., Owen C.B., Rivas A.A., Metz J.G., Apt K.E. Characterization of both polyunsaturated fatty acid biosynthetic pathways in Schizochytrium sp. Lipids. 2009;44:621–630. doi: 10.1007/s11745-009-3311-9. [DOI] [PubMed] [Google Scholar]
- 2.Cheng R.B., Lin X.Z., Wang Z.K., Yang S.J., Rong H., Ma Y. Establishment of a transgene expression system for the marine microalga Schizochytrium by 18S rDNA-targeted homologous recombination. World J Microb. Biot. 2011;27:737–741. doi: 10.1007/s11274-010-0510-8. [DOI] [Google Scholar]
- 3.Cheng R.B., Ma R., Li K., Rong H., Lin X.Z., Wang Z.K., Yang S.J., Ma Y. Agrobacterium tumefaciens mediated transformation of marine microalgae Schizochytrium. Microbiol. Res. 2012;167:179–186. doi: 10.1016/j.micres.2011.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Li Q., Zang X.N., Zhang X.C., Song X.J., Yang Q. Cloning and analysis of 18S rDNA gene from Schizochytrium limacinum OUC88 and 10 derived strains. Mar. Sci. 2014;38:71–78. [Google Scholar]
- 5.Steinberg N., Hamilton D. Bacteriophage P1 site-specific recombination. II. Recombination between loxP sites. J. Mol. Biol. 1981;150:467–486. doi: 10.1016/0022-2836(81)90375-2. [DOI] [PubMed] [Google Scholar]
- 6.Nafissi N., Slavcev R. Bacteriophage recombination systems and biotechnical applications. Appl. Biochem. Biotechnol. 2014;98:2841–2851. doi: 10.1007/s00253-014-5512-2. [DOI] [PubMed] [Google Scholar]
- 7.Guo F., Gopaul D.N., van Duyne G.D. Structure of Cre recombinase complexed with DNA in a site-specific recombination synapse. Nature. 1997;389:40–46. doi: 10.1038/37925. [DOI] [PubMed] [Google Scholar]
- 8.Dale E.C., Ow D.W. Intra-and intermolecular site-specific recombination in plant cells mediated by bacteriophage P1 recombinase. Gene. 1990;91:79–85. doi: 10.1016/0378-1119(90)90165-N. [DOI] [PubMed] [Google Scholar]
- 9.Vega J.M., Yu W.C., Han F.Q., Kato A., Peters E.M., Zhang Z.Y.J., Birchler J.A. Agrobacterium-mediated transformation of maize (Zea mays) with Cre-lox site specific recombination cassettes in BIBAC vectors. Plant Mol. Biol. 2008;66:587–598. doi: 10.1007/s11103-007-9276-2. [DOI] [PubMed] [Google Scholar]
- 10.Khattri A., Nandy S., Srivastava V. Heat-inducible Cre-lox system for marker excision in transgenic rice. J. Biosci. 2011;36:37–42. doi: 10.1007/s12038-011-9010-8. [DOI] [PubMed] [Google Scholar]
- 11.Liang M.T., Yang C.P., Xie Z.P., Staehelin C. Use of the Cre-loxP recombination system as an estimate for Agrobacterium-mediated co-transformation of tobacco leaves. Biotechnol. Lett. 2012;34:747–754. doi: 10.1007/s10529-011-0810-6. [DOI] [PubMed] [Google Scholar]
- 12.Zou X.Q., Peng A.H., Xu L.Z., Liu X.F., Lei T.G., Yao L.X., He Y.R., Chen S.C. Efficient auto-excision of a selectable marker gene from transgenic citrus by combining the Cre/loxP system and ipt selection. Plant Cell Rep. 2013;32:1601–1613. doi: 10.1007/s00299-013-1470-x. [DOI] [PubMed] [Google Scholar]
- 13.Kume S., Katayama N., Ichida K., Hattori-Ihara S., Nagasawa K., Yoshizaki G. Transgene manipulation in rainbow trout using Cre recombinase. Fish. Sci. 2014;80:767–773. doi: 10.1007/s12562-014-0742-x. [DOI] [Google Scholar]
- 14.Yu X.J., Chi Z., Wang F., Li J., Chi Z.M., Madzak C. Expression of the Acid Protease Gene from Saccharomycopsis fibuligera in the Marine-Derived Yarrowia lipolytica for Both Milk Clotting and Single Cell Protein Production. Appl. Biochem. Biotechnol. 2013;169:1993–2003. doi: 10.1007/s12010-013-0118-1. [DOI] [PubMed] [Google Scholar]
- 15.Guldener U., Heinisch J., Koehler G.J., Voss D., Hegemann J.H. A second set of loxP marker cassettes for Cre-mediated multiple gene knockouts in budding yeast. Nucleic Acids Res. 2002;30:e23. doi: 10.1093/nar/30.6.e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li Q., Zang X.N., Zhang X.C., Xu Y., Lu N. Sensitivities of Schizochytrium limacinum to seven antibiotics. J. Wuhan Univ. Nat. Sci. Ed. 2012;58:275–280. [Google Scholar]
- 17.Tian B.Y., Xu Y., Cai W.L., Huang Q., Gao Y.Y., Li X. Molecular cloning and overexpression of an endo-β-1,4-xylanase gene from Aspergillus niger in industrial Saccharomyces cerevisiae YS2 strain. Appl. Biochem. Biotechnol. 2013;170:320–328. doi: 10.1007/s12010-013-0173-7. [DOI] [PubMed] [Google Scholar]
- 18.Viau S., Maire M.A., Pasquis B., Grégoire S., Acar N., Bron A.M., Bretillon L., Creuzot-Garcher C.P., Joffre C. Efficacy of a 2-month dietary supplementation with polyunsaturated fatty acids in dry eye induced by scopolamine in a rat model. Graef. Arch. Clin. Exp. 2009;247:1039–1050. doi: 10.1007/s00417-009-1080-z. [DOI] [PubMed] [Google Scholar]
- 19.Diau G.Y., Hsieh A.T., Sarkadi-Nagy E.A., Wijendran V., Nathanielsz P.W., Brenna J.T. The influence of long chain polyunsaturate supplementation on docosahexaenoic acid and arachidonic acid in baboon neonate central nervous system. BMC Med. 2005;3:11. doi: 10.1186/1741-7015-3-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sun H.J., Meng X.Z., Han J.H., Zhang Z., Wang B., Bai X.D., Zhang X. Anti-cancer activity of DHA on gastric cancer: An in vitro and in vivo study. Tumor Biol. 2013;34:3791–3800. doi: 10.1007/s13277-013-0963-0. [DOI] [PubMed] [Google Scholar]
- 21.Robinson L.E., Mazurak V.C. N-3 Polyunsaturated Fatty Acids: Relationship to Inflammation in Healthy Adults and Adults Exhibiting Features of Metabolic Syndrome. Lipids. 2013;48:319–332. doi: 10.1007/s11745-013-3774-6. [DOI] [PubMed] [Google Scholar]
- 22.Lorente-Cebrián S., Costa A.G.V., Navas-Carretero S., Zabala M., Martínez J.A., Moreno-Aliaga M.J. Role of omega-3 fatty acids in obesity, metabolic syndrome, and cardiovascular diseases: A review of the evidence. J. Physiol. Biochem. 2013;69:633–651. doi: 10.1007/s13105-013-0265-4. [DOI] [PubMed] [Google Scholar]
- 23.Cheng R.B., Ge Y.Q., Yang B., Zhong X.M., Lin X.Z., Huang Z. Cloning and functional analysis of putative malonyl-CoA: Acyl-carrier protein transacylase gene from the docosahexaenoic acid-producer Schizochytrium sp. TIO1101. World J. Microb. Biot. 2013;29:959–967. doi: 10.1007/s11274-013-1253-0. [DOI] [PubMed] [Google Scholar]
- 24.Yan J.F., Cheng R.B., Lin X.Z., You S., Li K., Rong H., Ma Y. Overexpression of acetyl-CoA synthetase increased the biomass and fatty acid proportion in microalga Schizochytrium. Appl. Microbiol. Biotechnol. 2013;97:1933–1939. doi: 10.1007/s00253-012-4481-6. [DOI] [PubMed] [Google Scholar]
- 25.Zhu L., Zhang X., Chang L., Wang A., Feng P., Han L. Molecular cloning, prokaryotic expression and promoter analysis of squalene synthase gene from Schizochytrium Limacinum. Appl. Biochem. Microbiol. 2014;50:411–419. doi: 10.1134/S0003683814040140. [DOI] [Google Scholar]