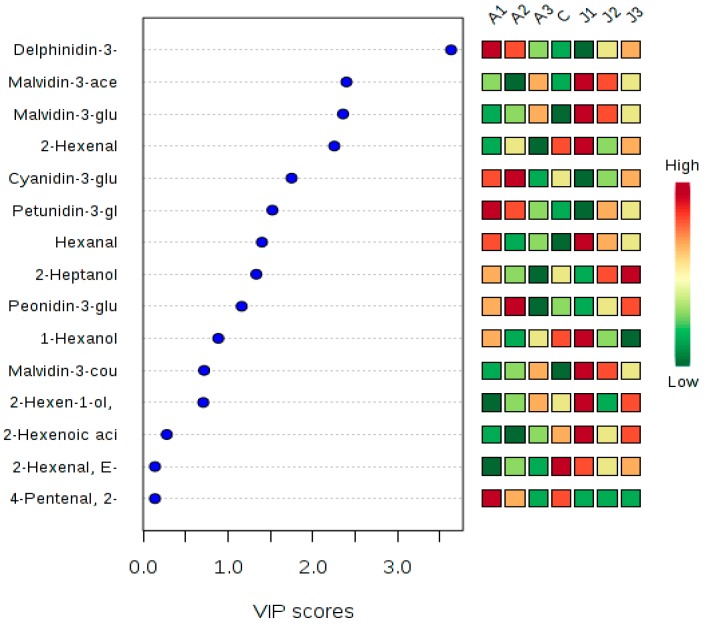
Figure 3.
Important features identified by PLS-DA. Partial Least Squares-Discriminant Analysis (PLS-DA) model was built between the data (X) and the permuted class labels (Y) using the optimal number of components determined by cross validation for the model based on the original class assignment. Variable Importance in Projection (VIP) is a weighted sum of squares of the PLS loadings taking into account the amount of explained Y-variation in each dimension. VIP scores are calculated for each component. The colored boxes on the right indicate the relative concentration of the corresponding metabolites in each group under study. A1: treatment with 1000 mg·L−1 ABA; A2: treatment with 600 mg·L−1 ABA; A3: treatment with 200 mg·L−1 ABA; J1: treatment with 800 µmol·L−1 MeJA; J2: treatment with 200 µmol·L−1 MeJA; J3: treatment with 50 µmol·L−1 MeJA; C: control. Values are the mean of three replicates.