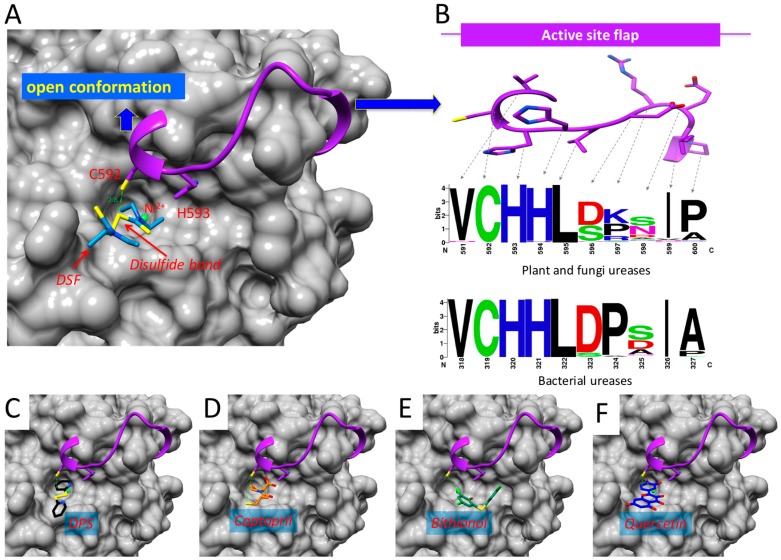
Figure 5.
Binding model of DSF and other reactive compounds to urease. (A) Model of the binding of DSF to JBU enzyme showing the interaction with Cys592 (dashed line, distance in Å). Ni2+ ions are shown in ball representations, active site flexible flap is shown in cartoon representations with Cys592 and His593 depicted in stick models and the rest of JBU is in surface representation. (B) Frequency of residues present in the active site flexible flap. The multiple alignment of urease enzymes available in Data Bank was used to plot frequencies in the logo. The amino acid residues are depicted as color code on the basis of their physicochemical properties as stated default it the weblogo server (http://weblogo.berkeley.edu/logo.cgi: polar amino acids (G,S,T,Y,C,Q,N) are green, basic (K,R,H) blue, acidic (D,E) red and hydrophobic (A,V,L,I,P,W,F,M) are black). (C) Model of the binding of DPS (D) Captopril; (E) Bithionol and (F) Quercetin to JBU. All structural images were generated using USCF-Chimera and are represented in a similar way to A.