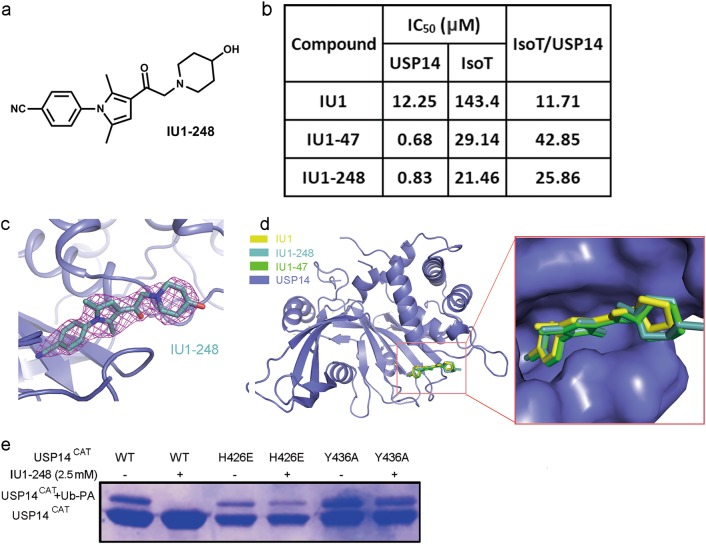
Fig. 4.
Structural-guided inhibitor design. a Chemical structure of IU1-248. b A table of the IC50 values of IU1 and its derivatives IU1-248 and IU1-47 toward proteasome-bound USP14 and IsoT. Similar to IU1-47, IU1-248 exhibited more than 10-fold higher potency and better selectivity than IU1. c 2|Fo| − |Fc| electron density maps, contoured at 1.5 σ and covering almost all the atoms of IU1-248. d Structural alignment of USP14CAT-IU1, USP14CAT-IU1-248 and USP14CAT-IU1-47 demonstrated that these inhibitors shared an identical binding pocket. e H426 and Y436 are involved in IU1-248 recognition, as proved by the Ub-PA assay. The H426E or Y436A mutation rescued the Ub-PA covalent binding activity of USP14 upon the addition of 2.5 mM IU1-248