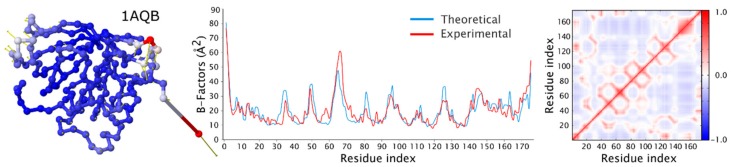
Figure 6.
Evaluation of fluctuational dynamics of retinol binding protein (PDB code: 1aqb, chain A) using DynOmics server. First panel shows protein structure with residues represented by balls located at the positions of Cα atoms and colored according to the values of B-factors (with red representing highly mobile and blue low mobile residues). Yellow arrows show the direction of oscillation. The chart in the middle compares experimental B-factors (in red) with predicted theoretical values (in blue) as a function of residue index. The effective force constant of the GNM springs is 0.92 kBTÅ−2, and corresponding rescaling prefactor is 85.40. Chart on the right shows cross-correlations (CC) between residue fluctuations with the coloring scheme shown on the right (with red denoting highly correlated and blue highly anticorrelated motions).