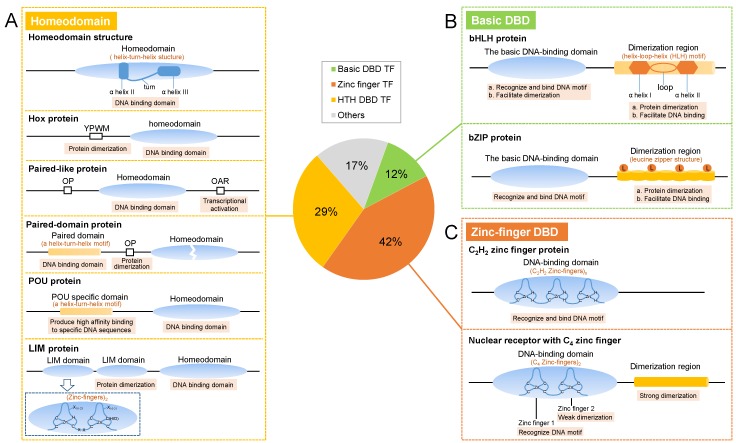
Figure 2.
The main structures of insect TFs. The pie chart represents the statistical ratio of the Drosophila TFs in different superclasses based on Hammonds’ study [22]. (A) Structures of homeodomain TFs. Homeodomain: a typical homeodomain contains a short N-terminal arm facilitating DNA binding and four α-helices with a helix II-turn-helix III structure that is responsible for DNA binding and recognition; Hox protein: in addition to the homeodomain, a Hox protein typically contains a YPWM motif in the N-terminus mediating protein dimerization; Paired-like protein: in addition to the homeodomain, some paired-like proteins include an N-terminal octapeptide (OP) motif, and some contain a C-terminal OAR motif that can be involved in transcriptional activation; Paired-domain protein: paired-domain proteins carry an N-terminal paired box with a helix-turn-helix (HTH) structure that mediates DNA binding, and some proteins have a full-length or truncated homeodomain in the C-terminus, as well as an OP motif between the paired box and the homeodomain to mediate protein dimerization; POU (Pit-Oct-Unc) protein: POU proteins have a POU-specific domain with an HTH structure in the N-terminus that contributes to the generation of high-affinity DNA binding, and the homeodomain in the C-terminus is responsible for DNA binding; LIM protein: LIM proteins possess two LIM domains with zinc finger (ZF) structures upstream of the homeodomain that mainly mediate protein-protein interactions. (B) Structures of basic DBD TFs. The basic leucine zipper (bZIP) protein: bZIP proteins harbor a basic DNA-binding domain (DBD) for specific DNA recognition and binding, and a leucine zipper domain in the C-terminus for protein dimerization and DNA binding. The leucine zipper domain forms dextrorotatory α-helixes, and a leucine appears in the seventh position of every seven amino acids; thus, an adjacent leucine appears every two turns on the same side of the helix. The basic helix-loop-helix (bHLH) protein: bHLH proteins are composed of a basic DBD in the N-terminus, followed by an HLH domain. The basic DBD accounts for DNA motif recognition and binding and facilitates protein dimerization. In the HLH domain, two hydrophilic and lipophilic α-helices are separated by a loop to form an HLH structure mediating protein dimerization and contributing to DNA binding. (C) Structures of the ZF TFs. C2H2 ZF protein: the C2H2 ZF protein has multiple connected ZF DBDs. In the root of every ZF, two cysteines and two histidines link Zn2+ to form a tetrahedron. Nuclear receptor (NR) with C4 ZF: NR contains a C4-type ZF region as a DBD that consists of eight conserved cysteine residues coordinated with two Zn2+ to form two ZFs with a tetrahedral coordination structure. The first ZF provides DNA-binding specificity, and the second ZF has a weak dimerization interface, allowing for dimerization of the receptor molecule. In addition, a ligand-binding domain is typically found in the C-terminus and functions as the main dimerization region.