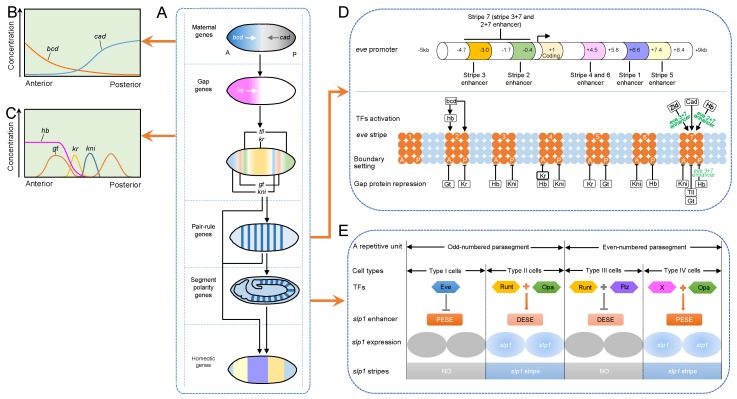
Figure 3.
Establishment of the embryonic anterior–posterior (A-P) axis in Drosophila. The diagram is adapted from Gilbert’s works [40,41]. (A) Regulatory hierarchy of the formation of Drosophila A-P axis patterning. The maternal effect proteins Bicoid (Bcd) and Caudal (Cad) form a concentration gradient along the A-P axis and generate specific positional information to activate the expression of the gap gene hunchback (Hb). Hh further initiates the proper expression of other gap genes along the A-P axis. Gap proteins subsequently activate the expression of pair-rule genes, which form seven stripes perpendicular to the A-P axis and divide those discontinuous regions defined by the gap gene into body segments. The pair-rule proteins then regulate the expression of segment polarity genes in specific cells of each somite, and their 14 expression stripes establish the boundaries of parasegments. Finally, each segment is characterized by specifically expressed Hox genes. (B) The concentration gradient of the maternal effect proteins Bcd and Cad along the A-P axis in the early cleavage embryo. (C) The concentration changes in gap genes along the A-P axis. (D) Regulation of expression of the pair-rule gene even-skipped (eve) in seven stripes. The above region represents a partial promoter of the eve gene that contains five different enhancers responsible for the distinct stripes. The lower part illustrates how TFs regulate eve expression in different stripes. The black box represents the TF, the green characters indicate the enhancer of eve, and the orange circles display the cells expressing eve. The vertical bars with the letters “A” and “P” denote the anterior and posterior boundaries of the eve stripe, respectively, and the numbers show the number of eve bands. (E) Regulation of a repetitive unit of sloppy paired 1 (slp1) stripe [42]. The 14-stripe patterning constitutes seven repetitive units, each containing odd-numbered and even-numbered parasegments. The odd-numbered parasegment consists of two types of cells: two type I cells in the posterior half do not express slp1 and two type II cells in the posterior half that express slp1. The even-numbered parasegment also contains two types of cells: type III cells that do not express slp1 in the first half and the latter type IV cells expressing slp1. The expression of slp1 in different cell types is regulated by different pair-rule proteins in a specific combination to regulate the proximal early stripe element (PESE) or the distal early stripe element (DESE) of slp1. The colored hexagons indicate the pair-rule proteins, the orange quadrants represent the enhancer, the gray ovals exhibit the cells that do not express slp1, the blue ovals denote the cells expressing slp1, the gray rectangles with “No” show no slp1 strips, and the blue rectangles represent slp1 strips. TF abbreviations: Hb: Hunchback; Gt: Giant; Kr: Krüppel; Kni: Knirps; Tll: Tailess; Zld: Zelda; Opa: Odd-paired; Ftz: Fushi tarazu; X: represents an as yet unidentified Factor X.