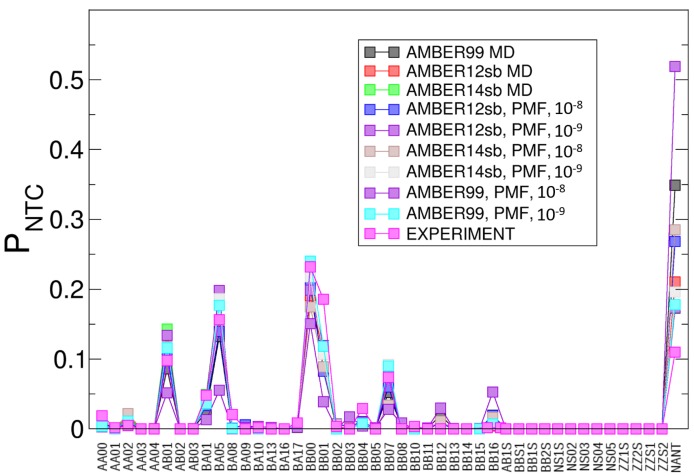
Figure 2.
Probability P for the occurrence of a structural class in the 6 different PMF-enriched, 3 MD simulations (AMBER99, AMBER12sb, AMBER14sb, with 2 coupling factors : and ), and a set of 29 experimental X-ray structures of the same dodecamer as a function of the class index. We used this analysis as the validation of the assignment of conformers and the technique using the auxiliary Hamiltonian in combination with a standard force field (the Hamiltonian ). The experimental values for these conformers equal 15.6 (BA05), 23.2 (BB00), and 7.3% (BB07), which is in good agreement with the simulation result. Only for the non-assigned class (NANT) do we observe a larger variation in values, ranging from 10 to above 50%, where the experimental value equals 11%. We conclude that the populations of the NtC classes from the PMF-enriched simulations are in good agreement with the MD simulations and the experimental PDB structures of the same molecule, which underlines a key property of the method, namely, the increase in the accessible conformation space of DNA toward the partition described in the PDB.