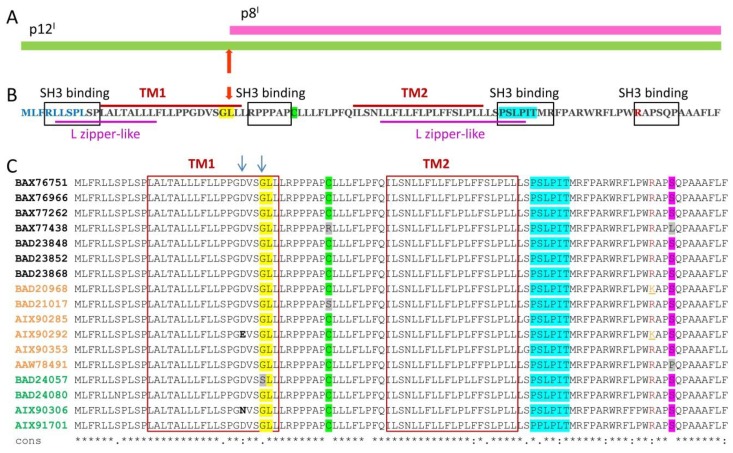
Figure 1.
p12I and p8I proteins’ organization: (A) p8I is a proteolytic product of 12I; the proteolytic cleavage site G29/L30 is indicated with an arrow; (B) aa sequence and putative domain architecture of full length p12I are shown: The endoplasmic reticulum (ER) retention N-terminal sequence is in blue; The transmembrane helices TM1 and TM2 are designated with red bars above the sequence; SH3 binding motifs are in black rectangles; L zipper-like motifs are underlined in magenta; R88 is in red; the G29/L30 cleavage site is highlighted in yellow and indicated by a red arrow. (C) Alignment of multiple aa sequences of p12I from randomly selected HTLV-1 strains, which were isolated from human carriers: Protein identification numbers in black, orange and green are from patients with adult T-cell leukemia/lymphoma (ATLL), patients with HTLV-1-associated myelopathy/tropical spastic paraparesis (HAM/TSP) and asymptomatic carriers, respectively. Conserved aa are indicated with asterisks under the sequences; residues D26 and G29 and their substitutions that are relevant to the level of p12I and p8I co-expression, are indicated with arrows; the cleavage site G29/L30, C39, the calcineurin binding motif, and residue S91, which is frequently mutated to other aa (mostly to P91), are highlighted in yellow, green blue, and magenta respectively; TM1 and TM2 are in red rectangles. The substitutions G29S, C39S, C39R, S91L, and S91P are highlighted in grey; R88 and K88 are in red and orange, respectively. T-COFFEE Multiple Sequence Alignment software was used.