Abstract
Peroxisome proliferator-activated receptor-delta (PPAR-δ), one of three members of the PPAR group in the nuclear receptor superfamily, is a ligand-activated transcription factor. PPAR-δ regulates important cellular metabolic functions that contribute to maintaining energy balance. PPAR-δ is especially important in regulating fatty acid uptake, transport, and β-oxidation as well as insulin secretion and sensitivity. These salutary PPAR-δ functions in normal cells are thought to protect against metabolic-syndrome-related diseases, such as obesity, dyslipidemia, insulin resistance/type 2 diabetes, hepatosteatosis, and atherosclerosis. Given the high clinical burden these diseases pose, highly selective synthetic activating ligands of PPAR-δ were developed as potential preventive/therapeutic agents. Some of these compounds showed some efficacy in clinical trials focused on metabolic-syndrome-related conditions. However, the clinical development of PPAR-δ agonists was halted because various lines of evidence demonstrated that cancer cells upregulated PPAR-δ expression/activity as a defense mechanism against nutritional deprivation and energy stresses, improving their survival and promoting cancer progression. This review discusses the complex relationship between PPAR-δ in health and disease and highlights our current knowledge regarding the different roles that PPAR-δ plays in metabolism, inflammation, and cancer.
Keywords: PPAR-δ, β-oxidation metabolism, inflammation, cancer
1. Introduction
Peroxisome proliferator-activated receptor-delta (PPAR-δ, also known as PPAR-β) is a member of the PPAR subgroup in the nuclear receptor superfamily. PPARs act as ligand-activated transcription factors that regulate important cellular metabolic functions [1]. Although PPAR-δ is ubiquitously expressed, its expression level in different tissues varies depending on cell type and disease status [2,3,4,5]. Homozygous knockout of murine Ppard through constructs targeting exon 4, which codes for the DNA binding domain, leads to embryonic lethality or impaired growth, which indicates that PPAR-δ plays a fundamental role in embryo development [6,7].
Details of PPAR structure and signaling mechanisms have been reviewed in detail in Reference [8] and will only be discussed briefly here. The characteristics of PPAR ligand-binding domains (LBD) allow for interaction of a broad range of potential ligands, including many lipid and lipid-like molecules [8]. Natural ligands for PPAR-δ include polyunsaturated fatty acids (PUFA, e.g., arachidonic and linoleic acid)) and their metabolites (e.g., prostacyclin/PGI2, 13S-hydroxyoctadecadienoic acid (13S-HODE), and 15S-hydroxyeicosatetraenoic acid (15S-HETE)) [9,10,11,12]. Although PPAR-δ has a narrower LBD relative to PPARs-α and-γ, binding pocket characteristics allow potential interaction with a variety of ligands, albeit many appear to bind at relatively low affinities [13]. While many potential endogenous ligands have been suggested in the literature, there is still some uncertainty about the physiological significance [14]. Selective ligands targeting PPAR-δ have also been developed, although none have been approved for clinical use to date [14,15].
PPAR signaling can be regulated in multiple ways, with outcomes depending upon whether PPAR and its binding partners are bound by ligand or not, ligand type (agonist, antagonist, partial agonist, etc.), and concentration as well as the availability of various coactivators or repressors [8]. The delivery of natural PPAR-δ ligands is facilitated by fatty acid transport proteins (FATPs) and fatty acid translocase (FAT, also known as CD36), which aid in import of extracellular lipids into the cell [16,17] and fatty-acid-binding proteins (FABPs), which transport cytoplasmic lipids within the cell [18,19]. Although most FABPs can bind a number of different lipids, it is unknown whether there is any selectivity in terms of the ligands FABP shuttles to PPARs [18,20,21]. In relation to PPAR-δ, FABP5 (also known as K-FABP or E-FABP) appears to be important for transport of lipid ligands to the nucleus [22]. Interestingly, FABP5 expression largely parallels that of PPAR-δ, and interaction between the two appears to be important in both normal and disease states, including many cancers [19]. Although a more detailed discussion is beyond the scope of this review, the interrelationship between the PPARs, their endogenous ligands, and various lipid transport proteins is complex, and several of these transport proteins are known transcriptional targets of PPARs (reviewed in References [16,19]).
Activation of PPARs by their ligands has been discussed in detail elsewhere and will be described only briefly here [8,23,24]. PPAR-δ activation requires interaction with various partners in the nucleus to transcriptionally regulate gene expression. Like other PPARs, PPAR-δ heterodimerizes with the retinoid X receptor (RXR) to activate or repress expression of downstream target genes by binding to PPAR response elements (PPREs) in their promoters [25,26]. In the absence of ligand binding, PPAR-RXR complexes are associated with corepressive factors and histone deacetylases that prevent transcriptional activation. Binding of an activating ligand to PPAR-δ leads to conformational changes that release corepressors and allow binding of coactivators [8,27]. In addition, PPARs can also engage in transrepression of other transcription factors. For example, in its unliganded state, PPAR-δ has been shown to form a complex with the transcription factor BCL-6, which prevents BCL-6 from repressing proinflammatory cytokine genes; therefore, this interaction promotes inflammation [28,29]. Conversely, binding of PPAR-δ agonist leads to disruption of the complex, and BCL-6 is freed to repress gene expression [28]. PPAR-δ has also been reported to interact with other transcription factors, such as β-catenin or NF-κB, to regulate gene expression [30,31].
Accumulating evidence has demonstrated that PPAR-δ can have distinct roles depending on the context (e.g., healthy vs. diseased, specific type of disease). While PPAR-δ allows normal cells (e.g., muscle cells and pancreatic cells) to better cope with adverse nutrient and energy pressures, PPAR-δ overexpression or hyperactivation can lead to promotion of inflammation and tumorigenesis. We will address some of the known discrepancies concerning PPAR-δ’s putative roles in metabolism, inflammation, and cancer in this review.
2. Metabolic Regulation by PPAR-δ
Modulation of cellular energy consumption is a major function of PPAR-δ. In muscle cells, ligand activation of PPAR-δ switches energy production from glycolysis to fatty acid oxidation as an alternative energy source, which can enhance muscle endurance [32]. In skeletal muscle cells, PPAR-δ activation by fatty acids increases fatty acid uptake and catabolism via β-oxidation [33]. Genetically targeting PPAR-δ overexpression in skeletal muscle cells increases succinate-dehydrogenase-positive muscle fibers with enhanced fatty acid oxidative capabilities and leads to an overall decrease in body fat [34]. Treatment of insulin-resistant obese monkeys with the PPAR-δ ligand GW501516 increased serum high-density lipoprotein cholesterol while decreasing low density lipoprotein, fasting triglycerides, and insulin [35]. Similarly, in vivo and in vitro transcriptome analyses of rodent muscle showed that PPAR-δ regulated downstream target genes required for fatty acid transport, β-oxidation of fatty acid, and mitochondrial respiration. Transgenic activation of PPAR-δ in mouse adipose tissues upregulated the expression of genes involved in fatty acid β-oxidation and energy dissipation via uncoupling of fatty acid oxidation and ATP production; these effects were observed in both ob/ob (mice homozygous for the spontaneous obese mutation (ob) in the leptin (Lep) gene) and wild-type (WT) mice on high-fat diet [36]. In contrast, PPAR-δ knockout mice were more prone to high-fat-diet-induced obesity [36]. A recent report showed induction of fatty acid oxidation in intestinal stem cells after a 24h fast; this effect was observed in both young and old mice and was mediated by the PPAR-δ target gene carnitine palmitoyltransferase 1 (Cpt1a) [37].
In addition to its effects on fatty acid oxidation, PPAR-δ leads to improved blood glucose homeostasis through a number of mechanisms. PPAR-δ is strongly expressed in pancreatic islet beta cells, promoting insulin secretion [38,39]. It is also protective against insulin resistance through effects on hepatic and peripheral energy substrate utilization [40,41,42]. Treatment of ob/ob mice, which have a genetic predisposition to obesity and diabetes, with GW50516 attenuated the ability of high-fat diet to induce obesity and insulin resistance and improved diabetes [43].
These salutary PPAR-δ functions in normal cells are thought to protect against metabolic-syndrome-related diseases, such as obesity, dyslipidemia, insulin resistance, hepatosteatosis, and atherosclerosis [44,45]. Therefore, highly selective synthetic PPAR-δ agonists (e.g., GW0742 [46], GW501516 [35]) were developed and tested clinically. However, improving cellular tolerance to an inhospitable metabolic microenvironment could also promote the survival of cancer cells (Figure 1). For example, overexpression of PPAR-δ was shown to improve breast cancer cell survival during low-glucose or hypoxic cell culture conditions through multiple mechanisms (e.g., enhanced antioxidant signaling, AKT/protein kinase B activation), and increased cell survival was inhibited with PPAR-δ antagonists [47]. Other studies have demonstrated that PPAR-δ promotion of fatty acid oxidation can lead to increased ATP production, contributing not only to the survival of breast cancer cells [48] but also other cancer cells, such as chronic lymphocytic leukemia cells [49]. Concerns regarding the potential protumorigenic effects of PPAR-δ have led to halting of the clinical development of PPAR-δ agonists [50,51].
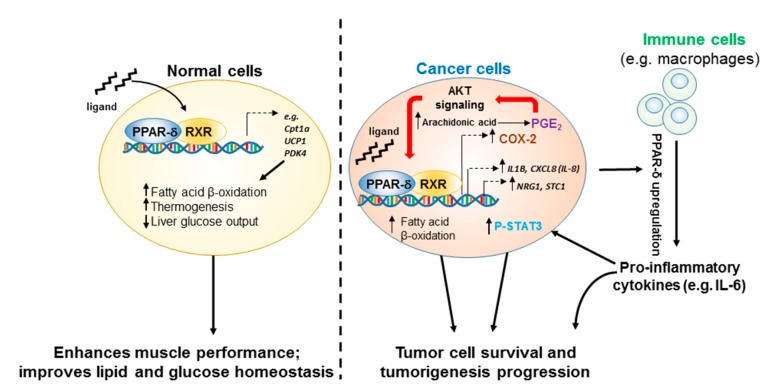
Figure 1.
Ligand-dependent actions of PPAR-δ in normal versus cancer cells. Binding of PPAR-δ agonists in normal cells (left) leads to the upregulation of genes associated with a switch to using fatty acids as an energy source (increased β-oxidation). It is also associated with systemic improvements in serum glucose regulation through effects on multiple tissues, including pancreas, adipose, liver, and muscle. In cancer cells (right), this capacity for PPAR-δ to promote use of fatty acid substrates as an energy source can enhance cell survival and proliferation under harsh metabolic conditions frequently found in tumors. In addition, both COX-2 and PI3K/AKT signaling pathways are often upregulated in tumor cells. Interaction of activated PPAR-δ with these key signaling hubs leads to establishment of a feed-forward circuit promoting cancer development and progression through upregulation of additional factors that enhance neoplastic processes in cancer cells themselves as well as noncancer cells (e.g., tumor-associated macrophages) that make up the tumor microenvironment. See text for additional details.
3. PPAR-δ in Inflammation-Related Diseases
Many studies have revealed that PPARs are involved in regulation of inflammation. Initially, PPARs were generally believed to have anti-inflammatory functions, and current research has more clearly defined such roles for PPAR-α and PPAR-γ [52,53]. PPAR-δ’s relationship with inflammation seems to be much different and still needs to be fully elucidated. In some contexts, PPAR-δ has been reported to have anti-inflammatory functions. For example, it was reported that the selective PPAR-δ agonist GW0742 alleviated inflammation in experimental autoimmune encephalomyelitis (EAE), while knockout of PPAR-δ aggravated EAE severity [54,55]. PPAR-δ’s antidiabetic functions also appear to be associated with reduced inflammatory signaling. In a rat model of type 2 diabetes, GW0742 was shown to reduce the proinflammatory cytokines tumor necrosis factor-α (TNF-α) and monocyte chemoattractant protein-1 (MCP-1) in liver tissues, in conjunction with reduced hepatic fat accumulation [56]. GW0742 was also shown to inhibit streptozotocin-induced diabetic nephropathy in mice through a reduction of inflammatory mediators, including MCP-1 and osteopontin [57]. In addition, a more recent study using both the db/db (homozygous for the spontaneous db mutation in the leptin receptor gene (Lepr)) and high-fat-diet-induced obese diabetic mouse models showed that PPAR-δ is a key mediator in exercise-induced reduction of vascular inflammation; PPAR-δ knockout mice did not exhibit this improvement with exercise [58].
In contrast to the reports mentioned above, PPAR-δ signaling appears to promote inflammation in other contexts. PPAR-δ expression is increased in patients with psoriasis, a common immune-mediated disease primarily affecting the skin [59]. In a transgenic mouse model, induction of PPAR-δ activation in the epidermis led to development of a psoriasis-like skin condition, which was correlated with increased IL-1 signaling and phosphorylation of STAT3 [59]. PPAR-δ signaling may also promote inflammation in some forms of arthritis. Mesenchymal stem cells (MSCs) have immunomodulatory properties that can limit inflammation [60]. In a collagen-induced mouse model of arthritis, mice receiving MSCs with reduced PPAR-δ activity (MSCs harvested from PPAR-δ knockout mice or WT PPAR-δ MSCs pretreated with the PPAR-δ antagonist GSK3787) had better suppression of inflammatory immune responses, leading to improvements in arthritis scores [61]. In the same study, inhibition of PPAR-δ with GSK3787 in human MSCs enhanced their ability to limit proliferation of peripheral blood mononuclear cells in coculture experiments [61].
4. Modulation of Inflammatory Actions in Immune Cells
Mechanistically, fatty acid oxidation is a central metabolic factor impacting immune cell differentiation and activation; for example, the behavior of inflammatory and immunosuppressive T cell and macrophage subsets can be affected by the balance of fatty acid oxidation and synthesis [62,63,64]. PPAR-δ’s roles as a fatty acid sensor and regulator of immune responses suggest a novel role for this receptor in metabolic modulation of immune cell differentiation and activity.
PPAR-δ appears to regulate inflammation through its effects on immune cells, particularly macrophages. Deletion of Ppard in liver-specific macrophages (Kupffer cells) led to reduced sensitivity to interleukin-4 and, therefore, impaired polarization to an alternative (M2-like) state with reduced inflammatory potential relative to M1-like macrophages [65]. This failure of differentiation to the M2 state ultimately led to hepatic dysfunction and insulin resistance, which was associated with altered fatty acid metabolism [65]. In another study using macrophage-specific PPAR-δ knockout mice, the deletion of Ppard was shown to decrease the phagocytosis of apoptotic cells and inhibit anti-inflammatory cytokine production by macrophages, leading to increased susceptibility to autoimmune kidney disease [66]. However, other evidence has revealed PPAR-δ’s modulation of inflammation may be even more complex. In primary human monocyte-derived macrophages (MDMs), PPAR-δ ligands were reported to repress inflammation-associated NF-κB and signal transducer and activator of transcription 1 (STAT1)-targeted genes (e.g., CXCL8 (IL-8) and CXCL1), yielding an M2-like macrophage phenotype [31]. Interestingly, they also observed reduced expression of factors associated with suppression of immune responses, such as indoleamine 2,3-dioxygenase 1 (IDO1), which plays an important role in limiting T-cell activation through its metabolism of tryptophan to kynurenine. In coculture experiments with MDMs and autologous T cells, MDMs exposed to PPAR-δ ligands increased CD8+/IFNγ+ T cell differentiation and limited IDO-1 mRNA and protein expression [31]. In addition, both kynurenine and the checkpoint inhibitor PD-1 ligand were also reduced in PPAR-δ ligand-treated MDMs [31]. Pathway analysis of RNA-Seq data from MDMs revealed upregulation of canonical PPAR-δ target genes involved in lipid metabolism, but exactly how they are engaged in immune regulation remains unclear [31]. Although the reports cited shed some light on the functions of PPAR-δ in immune cells, many questions remain concerning the mechanisms involved. The data reported above suggest that regulation of pro- and anti-inflammatory factor production and/or modulation of sensitivity to inflammatory stimuli may both be important in determining the overall outcome of PPAR-δ activation. The complexity of PPAR-δ’s effects on inflammation is likely to be related to its different modes of function as a transcription factor, which in turn are dependent on specific interactions with target gene sequences (PPREs) as well as the presence or absence of relevant PPAR-δ ligands and coactivators/repressors [67].
5. The Role of PPAR-δ in Cancer
5.1. PPAR-δ Crosstalk between Inflammation and Cancer
The role of chronic inflammation in promoting tumorigenesis is well recognized and considered to be a hallmark of cancer development [68,69]. Colitis-associated colon cancer (CAC) is one of best established examples of chronic inflammation’s role in increasing risk of cancer development, and its effects have been clearly demonstrated in preclinical and clinical studies [69,70]. PPAR-δ’s impact on inflammation-promoted tumorigenesis has been studied by various groups, especially in relation to lipid signaling, as in the case of prostaglandin E2 (PGE2) [71]. PGE2 is an eicosanoid lipid mediator generated through the actions of cyclooxygenases (COX-1 and -2). COX-2/ PGE2 signaling is frequently upregulated in tumors and is especially important in the context of inflammation-driven tumorigenesis; it has been particularly well-studied in the case of colonic tumorigenesis [72]. PGE2 enhanced PPAR-δ transcriptional activity via PI3 kinase/AKT activation to promote colon cancer cell survival in vitro and intestinal tumorigenesis in APCMin mice [73]. In addition, targeted overexpression of PPAR-δ in intestinal epithelial cells promoted development of azoxymethane (AOM)/dextran sodium sulfate (DSS)-induced CAC in mice via upregulation of IL-6/STAT3 [74]. Activation of PPAR-δ by the synthetic ligand GW501516 in colon cancer cell lines or primary mouse intestinal epithelial cells upregulated COX-2 expression and PGE2 production, subsequently increasing macrophage production of proinflammatory cytokines (e.g., CXCL1,CXCL2,CXCL4, and IL-1β) [75]. Both chemically and genetically induced colitis and CAC were markedly suppressed in a PPAR-δ knockout mouse model targeting PPAR-δ’s DNA binding domain (deleting exons 4 and 5), therefore blocking its function as a transcription factor [75]. Considered together, these findings suggest a positive feedback loop between PPAR-δ and COX-2 that orchestrates a proinflammatory microenvironment to enhance tumorigenesis. However, in studies using a different PPAR-δ knockout mouse model targeting the C-terminal portion of the protein (Ppard exon 8), female KO mice treated with DSS showed significant increases in some clinical colitis scores (weight loss and colon length) as well as levels of IFN-γ, TNF-α, IL-6, and worsened histopathological scores compared to WT mice [76,77]. These discordant data have been suggested to be secondary to the Ppard exon 8 genetic deletion strategy producing a hypomorphic PPAR-δ protein with some retained/altered function as opposed to a complete loss of function [75]. Interestingly, prostatic-epithelial-targeted genetic deletion of PPAR-δ’s DNA binding domain [7] has been recently reported to increase cellularity; in this setting, PPAR-δ was proposed to suppress prostate cancer via DNA-binding-dependent but ligand-binding-independent mechanisms [78].
PPAR-δ’s promotion of inflammation and tumorigenesis is not limited to the colon. Tumorigenesis repurposes various components of the inflammatory machinery to create a microenvironment conducive to tumor growth. For example, PPAR-δ is upregulated in tumor-associated macrophages (TAMs) in ovarian-cancer-associated ascites [79], and additional work has shown that macrophages associated with ovarian cancer tend to exhibit protumorigenic properties (e.g., immunosuppression, growth promotion) [80]. In a mouse model of breast cancer, mammary-epithelium-targeted PPAR-δ overexpression promotes tumorigenesis, which is further augmented by treatment with the PPAR-δ agonist GW501516 [81]. Tumor development in this model is associated with upregulation of proinflammatory genes, including COX-2, and activation of AKT signaling [81], reminiscent of the positive association between PPAR-δ, COX-2, and AKT signaling in colorectal tumorigenesis as discussed above. Likewise, a similar positive feedback loop between PPAR-δ and COX-2 signaling has also been demonstrated to enhance the survival of hepatocellular carcinoma cell lines [82].
Overall, while the genetic PPAR-δ deletion models generated via targeting exons encoding the PPAR-δ C-terminal region versus the DNA binding domain have yielded discordant data, the preponderance of evidence supports a model in which PPAR-δ strongly enforces inflammation-driven promotion of tumorigenesis through the enhancement of proinflammatory and protumorigenic mechanisms, especially as illustrated in the case of the positive feedback between PPAR-δ and COX-2 (Figure 1).
5.2. PPAR-δ Promotion of Cancer
While PPAR-δ’s relationship to cancer remains controversial [5,83], as more data continue to be published, we should see better clarification of PPAR-δ’s specific contributions to the tumorigenic process. PPAR-δ is overexpressed in various human cancers, including colorectal cancer (CRC) [84,85,86], where it can be upregulated even in early stages (e.g., in adenomas) [84]. Similarly, PPAR-δ is upregulated in other human malignancies, including pancreatic cancer, where its upregulation is correlated with higher pathological grade and increased risk of metastasis [87]. PPAR-δ is also known to be expressed in human lung cancer [88]. Of interest, while PPAR-δ protein expression as assessed by immunohistochemistry has only been observed in the nuclei of normal cells, it becomes nuclear and cytoplasmic in cancer cells [85,86]. The significance of this shift in PPAR-δ distribution in relation to its function is unknown. Generally, high PPAR-δ expression in human cancers is associated with negative survival outcomes [67,86].
The controversy regarding PPAR-δ’s role in tumorigenesis primarily stems from preclinical studies as illustrated by the following examples. First, a study using a PPAR-δ knockout mouse model (c-terminal KO/exon 8) showed that Ppard germline deletion increased the formation of colon tumors when the mice were bred with APCMin mice or treated with AOM [89]. Later, a study by another group showed opposite results; in this case, Ppard germline deletion (DNA binding domain KO/exons 4–5) reduced the formation of colon tumors when the mice were bred with APCMin mice [90]. Studies by our group showed that Ppard deletion genetically targeted to the intestinal epithelium profoundly inhibited AOM-induced colonic tumorigenesis [91]. While informative, the clinical relevance of using PPAR-δ knockout models of colorectal tumorigenesis in which PPAR-δ expression is reduced to levels below constitutive levels in normal cells is limited because PPAR-δ is typically upregulated in human colorectal cancer [11,84,86,92]. Modeling PPAR-δ’s influence on CRC by targeting its overexpression to the intestinal epithelial cells better simulated its upregulation in human colon cancer tissues and allowed clear demonstration that PPAR-δ overexpression strongly promoted AOM-induced colorectal tumorigenesis [93].
Interestingly, when PPAR-δ knockout mice with the c-terminal (exon 8) deletion, which led to increased colonic tumorigenesis in the initial study [89], were backcrossed to MMTV-COX-2 transgenic mice on the FVB/N background, COX-2-induced mammary tumorigenesis was markedly suppressed [94]. Furthermore, subsequent studies in which syngeneic PPAR-δ-expressing B16-F10 melanoma cells or Lewis lung cancer cells were implanted into the same PPAR-δ knockout mouse model also showed inhibition of tumorigenesis [95]. These contradictory findings using the same PPAR-δ knockout construct have been interpreted as suggesting that PPAR-δ has different roles depending on where it is expressed—specifically, that PPAR-δ expressed in noncancerous cells in the tumor microenvironment promotes tumorigenesis, whereas PPAR-δ expressed in cancer cells suppresses tumorigenesis [95]. However, in experiments where B16-F10 mouse melanoma cells in which PPAR-δ was downregulated using shRNA were subcutaneously injected into syngeneic (C57BL/6) WT or PPAR-δ germline knockout mice [74], PPAR-δ downregulation in either cancer or noncancer cells inhibited metastasis, although this effect was stronger when PPARδ expression was suppressed in cancer cells [86]. In addition, downregulation of PPAR-δ expression in cancer cells strongly suppressed metastases in orthotopic injection mouse models of multiple human cancers (e.g., colon, lung, melanoma, breast, and pancreas) via downregulation of important prometastatic genes (e.g., NRG1, CXCL8 (encoding IL-8), and STC1) in cancer cells and suppression of critical metastatic events including angiogenesis, epithelial-mesenchymal transition (EMT), and cancer cell invasion and migration [86] (Figure 1). PPAR-δ upregulation in human colon, lung, and breast cancers is also correlated with reduced metastasis-free survival [86]. PPAR-δ has also been reported to promote progression of melanoma via Snail expression [96] and prostate cancer via tumorigenic redirection of transforming growth factor β1 (TGF-β1) signaling [97]. A recently published study using unbiased global transcriptome analysis identified PPAR-δ activation as a driver of intestinal stem cell transformation and tumor promotion in APCMin mice maintained on a high-fat diet, suggesting it may play a mechanistic role in obesity-driven cancers [98]. Overall, a scenario is emerging in which PPAR-δ significantly contributes to both the initiation and progression of tumorigenesis in multiple tissues/tumor types.
6. Conclusions
PPAR-δ is a transcription factor that profoundly influences important cellular functions regulating metabolism and inflammation. PPAR-δ’s ability to act as a metabolic switch, shifting cellular energy utilization from glycolysis to fatty acid β-oxidation and thereby improving systemic glycemic control and lipid metabolism, make it an attractive target for prevention or treatment of metabolic-syndrome-related diseases (e.g., obesity, dyslipidemia, diabetes). Nevertheless, emerging data suggest that these same PPAR-δ-triggered mechanisms that help normal cells endure environmental metabolic challenges can also be exploited by cancer cells to promote their survival and, ultimately, cancer progression (Figure 1). Therefore, future therapeutic agents targeting PPAR-δ activation to treat metabolic diseases must be carefully designed and evaluated for any potential risks and off-target effects, including unintended promotion of preneoplastic or neoplastic lesions. Furthermore, our current understanding of the roles of PPAR-δ’s interactions with its endogenous ligands, lipid transporters, and other nuclear receptors, coactivators, and repressors remains incomplete. While some exciting discoveries have been made, more studies are still needed to better define the roles and mechanistic actions of PPAR-δ in different physiological and pathophysiological conditions.
Abbreviations
| AKT | Cellular homolog of viral akt gene, also known as protein kinase B |
| AOM | azoxymethane |
| APC | Adenomatous polyposis coli |
| BCL-6 | B cell lymphoma 6 |
| CAC | colitis-associated cancer |
| CRC | Colorectal cancer |
| COX-2 | cyclooxygenase-2 |
| CXCL1 | Chemokine (C-X-C motif) ligand 1 |
| CXCL2 | Chemokine (C-X-C motif) ligand 2 |
| CXCL4 | Chemokine (C-X-C motif) ligand 4 |
| DSS | dextran sodium sulfate |
| EAE | experimental autoimmune encepaholmyelitis |
| EMT | Epithelial mesenchymal transition |
| FABP | Fatty acid binding protein |
| FAT | Fatty acid translocase |
| FATP | Fatty acid transport protein |
| IDO-1 | indoleamine 2,3-dioxygenase 1 |
| IFN-γ | Interferon-γ |
| IL-1β | Interleukin-1β |
| IL-6 | Interleukin-6 |
| IL-8 | Interleukin-8 |
| MCP | monocyte chemoattractive protein |
| MDM | monocyte-derived macrophage |
| MSC | mesenchymal stem cell |
| MMTV | Mouse mammary tumor virus |
| NRG1 | Neuregulin 1 |
| NF-κB | Nuclear factor kappa-light-chain-enhancer of activated B cells |
| PGE2 | prostaglandin E2 |
| PI3K | phosphatidyl inositol 3 kinase |
| PPRE | PPAR response element |
| PUFA | polyunsaturated fatty acid |
| RXR | retinoid X receptor |
| STAT1/3 | Signal Transducer and Activator of Transcription 1/3 |
| STC1 | Stanniocalcin 1 |
| TAM | tumor-associated macrophage |
| TGF-β1 | transforming growth factor-β 1 |
| TNF-α | tumor necrosis factor α |
Author Contributions
Conception and design: Y.L., J.K.C., and I.S. Conceptual feedback and writing of the manuscript: Y.L., J.K.C., X.Z., J.J., D.W., and I.S.
Funding
This work was supported by grants R01CA142969, R01CA195686, and R01CA206539 awarded to I.S. from the National Cancer Institute at the National Institutes of Health, and grant RP140224 from the Cancer Prevention and Research Institute of Texas (to I.S).
Conflicts of Interest
The authors declare no competing interests.
References
- 1.Wagner K.D., Wagner N. Peroxisome proliferator-activated receptor beta/delta (PPARbeta/delta) acts as regulator of metabolism linked to multiple cellular functions. Pharmacol. Ther. 2010;125:423–435. doi: 10.1016/j.pharmthera.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Braissant O., Foufelle F., Scotto C., Dauca M., Wahli W. Differential expression of peroxisome proliferator-activated receptors (PPARs): Tissue distribution of PPAR-alpha, -beta, and -gamma in the adult rat. Endocrinology. 1996;137:354–366. doi: 10.1210/endo.137.1.8536636. [DOI] [PubMed] [Google Scholar]
- 3.Kliewer S.A., Forman B.M., Blumberg B., Ong E.S., Borgmeyer U., Mangelsdorf D.J., Umesono K., Evans R.M. Differential expression and activation of a family of murine peroxisome proliferator-activated receptors. Proc. Natl. Acad. Sci. USA. 1994;91:7355–7359. doi: 10.1073/pnas.91.15.7355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Auboeuf D., Rieusset J., Fajas L., Vallier P., Frering V., Riou J.P., Staels B., Auwerx J., Laville M., Vidal H. Tissue distribution and quantification of the expression of mRNAs of peroxisome proliferator-activated receptors and liver X receptor-alpha in humans: No alteration in adipose tissue of obese and NIDDM patients. Diabetes. 1997;46:1319–1327. doi: 10.2337/diab.46.8.1319. [DOI] [PubMed] [Google Scholar]
- 5.Xu M., Zuo X., Shureiqi I. Targeting peroxisome proliferator-activated receptor-beta/delta in colon cancer: How to aim? Biochem. Pharmacol. 2013;85:607–611. doi: 10.1016/j.bcp.2012.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nadra K., Anghel S.I., Joye E., Tan N.S., Basu-Modak S., Trono D., Wahli W., Desvergne B. Differentiation of trophoblast giant cells and their metabolic functions are dependent on peroxisome proliferator-activated receptor beta/delta. Mol. Cell. Boil. 2006;26:3266–3281. doi: 10.1128/MCB.26.8.3266-3281.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barak Y., Liao D., He W., Ong E.S., Nelson M.C., Olefsky J.M., Boland R., Evans R.M. Effects of peroxisome proliferator-activated receptor delta on placentation, adiposity, and colorectal cancer. Proc. Natl. Acad. Sci. USA. 2002;99:303–308. doi: 10.1073/pnas.012610299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Harmon G.S., Lam M.T., Glass C.K. PPARs and lipid ligands in inflammation and metabolism. Chem. Rev. 2011;111:6321–6340. doi: 10.1021/cr2001355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Xu H.E., Lambert M.H., Montana V.G., Parks D.J., Blanchard S.G., Brown P.J., Sternbach D.D., Lehmann J.M., Wisely G.B., Willson T.M., et al. Molecular Recognition of Fatty Acids by Peroxisome Proliferator–Activated Receptors. Mol. Cell. 1999;3:397–403. doi: 10.1016/S1097-2765(00)80467-0. [DOI] [PubMed] [Google Scholar]
- 10.Shureiqi I., Jiang W., Zuo X., Wu Y., Stimmel J.B., Leesnitzer L.M., Morris J.S., Fan H.Z., Fischer S.M., Lippman S.M. The 15-lipoxygenase-1 product 13-S-hydroxyoctadecadienoic acid down-regulates PPAR-delta to induce apoptosis in colorectal cancer cells. Proc. Natl. Acad. Sci. USA. 2003;100:9968–9973. doi: 10.1073/pnas.1631086100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gupta R.A., Tan J., Krause W.F., Geraci M.W., Willson T.M., Dey S.K., DuBois R.N. Prostacyclin-mediated activation of peroxisome proliferator-activated receptor delta in colorectal cancer. Proc. Natl. Acad. Sci. USA. 2000;97:13275–13280. doi: 10.1073/pnas.97.24.13275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Naruhn S., Meissner W., Adhikary T., Kaddatz K., Klein T., Watzer B., Muller-Brusselbach S., Muller R. 15-hydroxyeicosatetraenoic acid is a preferential peroxisome proliferator-activated receptor beta/delta agonist. Mol. Pharmacol. 2010;77:171–184. doi: 10.1124/mol.109.060541. [DOI] [PubMed] [Google Scholar]
- 13.Xu H.E., Lambert M.H., Montana V.G., Plunket K.D., Moore L.B., Collins J.L., Oplinger J.A., Kliewer S.A., Gampe R.T., Jr., McKee D.D., et al. Structural determinants of ligand binding selectivity between the peroxisome proliferator-activated receptors. Proc. Natl. Acad. Sci. USA. 2001;98:13919–13924. doi: 10.1073/pnas.241410198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wu C.C., Baiga T.J., Downes M., La Clair J.J., Atkins A.R., Richard S.B., Fan W., Stockley-Noel T.A., Bowman M.E., Noel J.P., et al. Structural basis for specific ligation of the peroxisome proliferator-activated receptor delta. Proc. Natl. Acad. Sci. USA. 2017;114:E2563–E2570. doi: 10.1073/pnas.1621513114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tan C.K., Zhuang Y., Wahli W. Synthetic and natural Peroxisome Proliferator-Activated Receptor (PPAR) agonists as candidates for the therapy of the metabolic syndrome. Expert Opin. Ther. Targets. 2017;21:333–348. doi: 10.1080/14728222.2017.1280467. [DOI] [PubMed] [Google Scholar]
- 16.Glatz J.F., Luiken J.J., Bonen A. Membrane fatty acid transporters as regulators of lipid metabolism: Implications for metabolic disease. Physiol. Rev. 2010;90:367–417. doi: 10.1152/physrev.00003.2009. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura M.T., Yudell B.E., Loor J.J. Regulation of energy metabolism by long-chain fatty acids. Prog. Lipid Res. 2014;53:124–144. doi: 10.1016/j.plipres.2013.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Makowski L., Hotamisligil G.S. Fatty acid binding proteins—The evolutionary crossroads of inflammatory and metabolic responses. J. Nutr. 2004;134:2464S–2468S. doi: 10.1093/jn/134.9.2464S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Amiri M., Yousefnia S., Seyed Forootan F., Peymani M., Ghaedi K., Nasr Esfahani M.H. Diverse roles of fatty acid binding proteins (FABPs) in development and pathogenesis of cancers. Gene. 2018;676:171–183. doi: 10.1016/j.gene.2018.07.035. [DOI] [PubMed] [Google Scholar]
- 20.Tan N.S., Shaw N.S., Vinckenbosch N., Liu P., Yasmin R., Desvergne B., Wahli W., Noy N. Selective cooperation between fatty acid binding proteins and peroxisome proliferator-activated receptors in regulating transcription. Mol. Cell. Boil. 2002;22:5114–5127. doi: 10.1128/MCB.22.14.5114-5127.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tan N.S., Vazquez-Carrera M., Montagner A., Sng M.K., Guillou H., Wahli W. Transcriptional control of physiological and pathological processes by the nuclear receptor PPARbeta/delta. Prog. Lipid Res. 2016;64:98–122. doi: 10.1016/j.plipres.2016.09.001. [DOI] [PubMed] [Google Scholar]
- 22.Armstrong E.H., Goswami D., Griffin P.R., Noy N., Ortlund E.A. Structural basis for ligand regulation of the fatty acid-binding protein 5, peroxisome proliferator-activated receptor beta/delta (FABP5-PPARbeta/delta) signaling pathway. J. Biol. Chem. 2014;289:14941–14954. doi: 10.1074/jbc.M113.514646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zoete V., Grosdidier A., Michielin O. Peroxisome proliferator-activated receptor structures: Ligand specificity, molecular switch and interactions with regulators. Biochim. Biophys. Acta. 2007;1771:915–925. doi: 10.1016/j.bbalip.2007.01.007. [DOI] [PubMed] [Google Scholar]
- 24.Batista F.A., Trivella D.B., Bernardes A., Gratieri J., Oliveira P.S., Figueira A.C., Webb P., Polikarpov I. Structural insights into human peroxisome proliferator activated receptor delta (PPAR-delta) selective ligand binding. PLoS ONE. 2012;7:e33643. doi: 10.1371/journal.pone.0033643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Feige J.N., Gelman L., Tudor C., Engelborghs Y., Wahli W., Desvergne B. Fluorescence imaging reveals the nuclear behavior of peroxisome proliferator-activated receptor/retinoid X receptor heterodimers in the absence and presence of ligand. J. Biol. Chem. 2005;280:17880–17890. doi: 10.1074/jbc.M500786200. [DOI] [PubMed] [Google Scholar]
- 26.Matsusue K., Miyoshi A., Yamano S., Gonzalez F.J. Ligand-activated PPARbeta efficiently represses the induction of LXR-dependent promoter activity through competition with RXR. Mol. Cell. Endocrinol. 2006;256:23–33. doi: 10.1016/j.mce.2006.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Viswakarma N., Jia Y., Bai L., Vluggens A., Borensztajn J., Xu J., Reddy J.K. Coactivators in PPAR-Regulated Gene Expression. PPAR Res. 2010;2010 doi: 10.1155/2010/250126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee C.H., Chawla A., Urbiztondo N., Liao D., Boisvert W.A., Evans R.M., Curtiss L.K. Transcriptional repression of atherogenic inflammation: Modulation by PPARdelta. Science. 2003;302:453–457. doi: 10.1126/science.1087344. [DOI] [PubMed] [Google Scholar]
- 29.Chinetti-Gbaguidi G., Staels B. PPARbeta in macrophages and atherosclerosis. Biochimie. 2017;136:59–64. doi: 10.1016/j.biochi.2016.12.008. [DOI] [PubMed] [Google Scholar]
- 30.Scholtysek C., Katzenbeisser J., Fu H., Uderhardt S., Ipseiz N., Stoll C., Zaiss M.M., Stock M., Donhauser L., Bohm C., et al. PPARbeta/delta governs Wnt signaling and bone turnover. Nat. Med. 2013;19:608–613. doi: 10.1038/nm.3146. [DOI] [PubMed] [Google Scholar]
- 31.Adhikary T., Wortmann A., Schumann T., Finkernagel F., Lieber S., Roth K., Toth P.M., Diederich W.E., Nist A., Stiewe T., et al. The transcriptional PPARbeta/delta network in human macrophages defines a unique agonist-induced activation state. Nucleic Acids Res. 2015;43:5033–5051. doi: 10.1093/nar/gkv331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Fan W., Waizenegger W., Lin C.S., Sorrentino V., He M.X., Wall C.E., Li H., Liddle C., Yu R.T., Atkins A.R., et al. PPARdelta Promotes Running Endurance by Preserving Glucose. Cell Metab. 2017;25:1186–1193. doi: 10.1016/j.cmet.2017.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Holst D., Luquet S., Nogueira V., Kristiansen K., Leverve X., Grimaldi P.A. Nutritional regulation and role of peroxisome proliferator-activated receptor δ in fatty acid catabolism in skeletal muscle. Biochim. Biophys. Acta Mol. Cell Boil. Lipids. 2003;1633:43–50. doi: 10.1016/S1388-1981(03)00071-4. [DOI] [PubMed] [Google Scholar]
- 34.Luquet S., Lopez-Soriano J., Holst D., Fredenrich A., Melki J., Rassoulzadegan M., Grimaldi P.A. Peroxisome proliferator-activated receptor δ controls muscle development and oxidative capability. FASEB J. 2003;17:2299–2301. doi: 10.1096/fj.03-0269fje. [DOI] [PubMed] [Google Scholar]
- 35.Oliver W.R., Jr., Shenk J.L., Snaith M.R., Russell C.S., Plunket K.D., Bodkin N.L., Lewis M.C., Winegar D.A., Sznaidman M.L., Lambert M.H., et al. A selective peroxisome proliferator-activated receptor delta agonist promotes reverse cholesterol transport. Proc. Natl. Acad. Sci. USA. 2001;98:5306–5311. doi: 10.1073/pnas.091021198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y.-X., Lee C.-H., Tiep S., Yu R.T., Ham J., Kang H., Evans R.M. Peroxisome-Proliferator-Activated Receptor δ Activates Fat Metabolism to Prevent Obesity. Cell. 2003;113:159–170. doi: 10.1016/S0092-8674(03)00269-1. [DOI] [PubMed] [Google Scholar]
- 37.Mihaylova M.M., Cheng C.W., Cao A.Q., Tripathi S., Mana M.D., Bauer-Rowe K.E., Abu-Remaileh M., Clavain L., Erdemir A., Lewis C.A., et al. Fasting Activates Fatty Acid Oxidation to Enhance Intestinal Stem Cell Function during Homeostasis and Aging. Cell Stem Cell. 2018;22:769–778. doi: 10.1016/j.stem.2018.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ravnskjaer K., Frigerio F., Boergesen M., Nielsen T., Maechler P., Mandrup S. PPARdelta is a fatty acid sensor that enhances mitochondrial oxidation in insulin-secreting cells and protects against fatty acid-induced dysfunction. J. Lipid Res. 2010;51:1370–1379. doi: 10.1194/jlr.M001123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Iglesias J., Barg S., Vallois D., Lahiri S., Roger C., Yessoufou A., Pradevand S., McDonald A., Bonal C., Reimann F., et al. PPARbeta/delta affects pancreatic beta cell mass and insulin secretion in mice. J. Clin. Investig. 2012;122:4105–4117. doi: 10.1172/JCI42127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lee C.H., Olson P., Hevener A., Mehl I., Chong L.W., Olefsky J.M., Gonzalez F.J., Ham J., Kang H., Peters J.M., et al. PPARdelta regulates glucose metabolism and insulin sensitivity. Proc. Natl. Acad. Sci. USA. 2006;103:3444–3449. doi: 10.1073/pnas.0511253103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang T., Abbott M.J., Ahmadian M., Lopes A.B., Wang Y., Sul H.S. Desnutrin/ATGL activates PPARdelta to promote mitochondrial function for insulin secretion in islet beta cells. Cell Metab. 2013;18:883–895. doi: 10.1016/j.cmet.2013.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Doktorova M., Zwarts I., Zutphen T.V., Dijk T.H.V., Bloks V.W., Harkema L., Bruin A., Downes M., Evans R.M., Verkade H.J., et al. Intestinal PPARdelta protects against diet-induced obesity, insulin resistance and dyslipidemia. Sci. Rep. 2017;7:846. doi: 10.1038/s41598-017-00889-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tanaka T., Yamamoto J., Iwasaki S., Asaba H., Hamura H., Ikeda Y., Watanabe M., Magoori K., Ioka R.X., Tachibana K., et al. Activation of peroxisome proliferator-activated receptor δ induces fatty acid β-oxidation in skeletal muscle and attenuates metabolic syndrome. Proc. Natl. Acad. Sci. USA. 2003;100:15924–15929. doi: 10.1073/pnas.0306981100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen J., Montagner A., Tan N., Wahli W. Insights into the Role of PPARβ/δ in NAFLD. Int. J. Mol. Sci. 2018;19:1893. doi: 10.3390/ijms19071893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reilly S.M., Lee C.H. PPAR delta as a therapeutic target in metabolic disease. FEBS Lett. 2008;582:26–31. doi: 10.1016/j.febslet.2007.11.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sznaidman M.L., Haffner C.D., Maloney P.R., Fivush A., Chao E., Goreham D., Sierra M.L., LeGrumelec C., Xu H.E., Montana V.G., et al. Novel selective small molecule agonists for peroxisome proliferator-activated receptor delta (PPARdelta)—Synthesis and biological activity. Bioorg. Med. Chem. Lett. 2003;13:1517–1521. doi: 10.1016/S0960-894X(03)00207-5. [DOI] [PubMed] [Google Scholar]
- 47.Wang X., Wang G., Shi Y., Sun L., Gorczynski R., Li Y.J., Xu Z., Spaner D.E. PPAR-delta promotes survival of breast cancer cells in harsh metabolic conditions. Oncogenesis. 2016;5:e232. doi: 10.1038/oncsis.2016.41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Carracedo A., Weiss D., Leliaert A.K., Bhasin M., de Boer V.C.J., Laurent G., Adams A.C., Sundvall M., Song S.J., Ito K., et al. A metabolic prosurvival role for PML in breast cancer. J. Clin. Investig. 2012;122:3088–3100. doi: 10.1172/JCI62129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li Y.J., Sun L., Shi Y., Wang G., Wang X., Dunn S.E., Iorio C., Screaton R.A., Spaner D.E. PPAR-delta promotes survival of chronic lymphocytic leukemia cells in energetically unfavorable conditions. Leukemia. 2017;31:1905–1914. doi: 10.1038/leu.2016.395. [DOI] [PubMed] [Google Scholar]
- 50.Sahebkar A., Chew G.T., Watts G.F. New peroxisome proliferator-activated receptor agonists: Potential treatments for atherogenic dyslipidemia and non-alcoholic fatty liver disease. Expert Opin. Pharmacother. 2014;15:493–503. doi: 10.1517/14656566.2014.876992. [DOI] [PubMed] [Google Scholar]
- 51.Cox R.L. Rationally designed PPARdelta-specific agonists and their therapeutic potential for metabolic syndrome. Proc. Natl. Acad. Sci. USA. 2017;114:3284–3285. doi: 10.1073/pnas.1702084114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Daynes R.A., Jones D.C. Emerging roles of PPARs in inflammation and immunity. Nat. Rev. Immunol. 2002;2:748–759. doi: 10.1038/nri912. [DOI] [PubMed] [Google Scholar]
- 53.Choi J.M., Bothwell A.L. The nuclear receptor PPARs as important regulators of T-cell functions and autoimmune diseases. Mol. Cells. 2012;33:217–222. doi: 10.1007/s10059-012-2297-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kanakasabai S., Chearwae W., Walline C.C., Iams W., Adams S.M., Bright J.J. Peroxisome proliferator-activated receptor delta agonists inhibit T helper type 1 (Th1) and Th17 responses in experimental allergic encephalomyelitis. Immunology. 2010;130:572–588. doi: 10.1111/j.1365-2567.2010.03261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dunn S.E., Bhat R., Straus D.S., Sobel R.A., Axtell R., Johnson A., Nguyen K., Mukundan L., Moshkova M., Dugas J.C., et al. Peroxisome proliferator-activated receptor delta limits the expansion of pathogenic Th cells during central nervous system autoimmunity. J. Exp. Med. 2010;207:1599–1608. doi: 10.1084/jem.20091663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee M.Y., Choi R., Kim H.M., Cho E.J., Kim B.H., Choi Y.S., Naowaboot J., Lee E.Y., Yang Y.C., Shin J.Y., et al. Peroxisome proliferator-activated receptor delta agonist attenuates hepatic steatosis by anti-inflammatory mechanism. Exp. Mol. Med. 2012;44:578–585. doi: 10.3858/emm.2012.44.10.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Matsushita Y., Ogawa D., Wada J., Yamamoto N., Shikata K., Sato C., Tachibana H., Toyota N., Makino H. Activation of peroxisome proliferator-activated receptor delta inhibits streptozotocin-induced diabetic nephropathy through anti-inflammatory mechanisms in mice. Diabetes. 2011;60:960–968. doi: 10.2337/db10-1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cheang W.S., Wong W.T., Zhao L., Xu J., Wang L., Lau C.W., Chen Z.Y., Ma R.C., Xu A., Wang N., et al. PPARdelta Is Required for Exercise to Attenuate Endoplasmic Reticulum Stress and Endothelial Dysfunction in Diabetic Mice. Diabetes. 2017;66:519–528. doi: 10.2337/db15-1657. [DOI] [PubMed] [Google Scholar]
- 59.Romanowska M., Reilly L., Palmer C.N., Gustafsson M.C., Foerster J. Activation of PPARbeta/delta causes a psoriasis-like skin disease in vivo. PLoS ONE. 2010;5:e9701. doi: 10.1371/journal.pone.0009701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bernardo M.E., Fibbe W.E. Mesenchymal stromal cells: Sensors and switchers of inflammation. Cell Stem Cell. 2013;13:392–402. doi: 10.1016/j.stem.2013.09.006. [DOI] [PubMed] [Google Scholar]
- 61.Luz-Crawford P., Ipseiz N., Espinosa-Carrasco G., Caicedo A., Tejedor G., Toupet K., Loriau J., Scholtysek C., Stoll C., Khoury M., et al. PPARbeta/delta directs the therapeutic potential of mesenchymal stem cells in arthritis. Ann. Rheum. Dis. 2016;75:2166–2174. doi: 10.1136/annrheumdis-2015-208696. [DOI] [PubMed] [Google Scholar]
- 62.O’Neill L.A.J., Kishton R.J., Rathmell J. A guide to immunometabolism for immunologists. Nat. Rev. Immunol. 2016;16:553–565. doi: 10.1038/nri.2016.70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Andrejeva G., Rathmell J.C. Similarities and Distinctions of Cancer and Immune Metabolism in Inflammation and Tumors. Cell Metab. 2017;26:49–70. doi: 10.1016/j.cmet.2017.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gerriets V.A., Kishton R.J., Nichols A.G., Macintyre A.N., Inoue M., Ilkayeva O., Winter P.S., Liu X., Priyadharshini B., Slawinska M.E., et al. Metabolic programming and PDHK1 control CD4+ T cell subsets and inflammation. J. Clin. Investig. 2015;125:194–207. doi: 10.1172/JCI76012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Odegaard J.I., Ricardo-Gonzalez R.R., Red Eagle A., Vats D., Morel C.R., Goforth M.H., Subramanian V., Mukundan L., Ferrante A.W., Chawla A. Alternative M2 activation of Kupffer cells by PPARdelta ameliorates obesity-induced insulin resistance. Cell Metab. 2008;7:496–507. doi: 10.1016/j.cmet.2008.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mukundan L., Odegaard J.I., Morel C.R., Heredia J.E., Mwangi J.W., Ricardo-Gonzalez R.R., Goh Y.P., Eagle A.R., Dunn S.E., Awakuni J.U., et al. PPAR-delta senses and orchestrates clearance of apoptotic cells to promote tolerance. Nat. Med. 2009;15:1266–1272. doi: 10.1038/nm.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Muller R. PPARbeta/delta in human cancer. Biochimie. 2017;136:90–99. doi: 10.1016/j.biochi.2016.10.019. [DOI] [PubMed] [Google Scholar]
- 68.Hanahan D., Weinberg R.A. Hallmarks of Cancer: The Next Generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- 69.Jess T., Rungoe C., Peyrin-Biroulet L. Risk of colorectal cancer in patients with ulcerative colitis: A meta-analysis of population-based cohort studies. Clin. Gastroenterol. Hepatol. 2012;10:639–645. doi: 10.1016/j.cgh.2012.01.010. [DOI] [PubMed] [Google Scholar]
- 70.Terzić J., Grivennikov S., Karin E., Karin M. Inflammation and Colon Cancer. Gastroenterology. 2010;138:2101–2114. doi: 10.1053/j.gastro.2010.01.058. [DOI] [PubMed] [Google Scholar]
- 71.Wang D., DuBois R.N. PPARdelta and PGE2 signaling pathways communicate and connect inflammation to colorectal cancer. Inflamm. Cell Signal. 2014;1 doi: 10.14800/ics.338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Backlund M.G., Mann J.R., Dubois R.N. Mechanisms for the prevention of gastrointestinal cancer: The role of prostaglandin E2. Oncology. 2005;69:28–32. doi: 10.1159/000086629. [DOI] [PubMed] [Google Scholar]
- 73.Wang D., Wang H., Shi Q., Katkuri S., Walhi W., Desvergne B., Das S.K., Dey S.K., DuBois R.N. Prostaglandin E2 promotes colorectal adenoma growth via transactivation of the nuclear peroxisome proliferator-activated receptor δ. Cancer Cell. 2004;6:285–295. doi: 10.1016/j.ccr.2004.08.011. [DOI] [PubMed] [Google Scholar]
- 74.Mao F., Xu M., Zuo X., Yu J., Xu W., Moussalli M.J., Elias E., Li H.S., Watowich S.S., Shureiqi I. 15-Lipoxygenase-1 suppression of colitis-associated colon cancer through inhibition of the IL-6/STAT3 signaling pathway. FASEB J. 2015;29:2359–2370. doi: 10.1096/fj.14-264515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Wang D., Fu L., Ning W., Guo L., Sun X., Dey S.K., Chaturvedi R., Wilson K.T., DuBois R.N. Peroxisome proliferator-activated receptor delta promotes colonic inflammation and tumor growth. Proc. Natl. Acad. Sci. USA. 2014;111:7084–7089. doi: 10.1073/pnas.1324233111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Peters J.M., Lee S.S., Li W., Ward J.M., Gavrilova O., Everett C., Reitman M.L., Hudson L.D., Gonzalez F.J. Growth, adipose, brain, and skin alterations resulting from targeted disruption of the mouse peroxisome proliferator-activated receptor beta(delta) Mol. Cell. Boil. 2000;20:5119–5128. doi: 10.1128/MCB.20.14.5119-5128.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hollingshead H.E., Morimura K., Adachi M., Kennett M.J., Billin A.N., Willson T.M., Gonzalez F.J., Peters J.M. PPARbeta/delta protects against experimental colitis through a ligand-independent mechanism. Dig. Dis. Sci. 2007;52:2912–2919. doi: 10.1007/s10620-006-9644-9. [DOI] [PubMed] [Google Scholar]
- 78.Martín-Martín N., Zabala-Letona A., Fernández-Ruiz S., Arreal L., Camacho L., Castillo-Martin M., Cortazar A.R., Torrano V., Astobiza I., Zúñiga-García P., et al. PPARδ Elicits Ligand-Independent Repression of Trefoil Factor Family to Limit Prostate Cancer Growth. Cancer Res. 2018;78:399–409. doi: 10.1158/0008-5472.CAN-17-0908. [DOI] [PubMed] [Google Scholar]
- 79.Reinartz S., Finkernagel F., Adhikary T., Rohnalter V., Schumann T., Schober Y., Nockher W.A., Nist A., Stiewe T., Jansen J.M., et al. A transcriptome-based global map of signaling pathways in the ovarian cancer microenvironment associated with clinical outcome. Genome Biol. 2016;17:108. doi: 10.1186/s13059-016-0956-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Colvin E.K. Tumor-Associated Macrophages Contribute to Tumor Progression in Ovarian Cancer. Front. Oncol. 2014;4:137. doi: 10.3389/fonc.2014.00137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yuan H., Lu J., Xiao J., Upadhyay G., Umans R., Kallakury B., Yin Y., Fant M.E., Kopelovich L., Glazer R.I. PPARdelta induces estrogen receptor-positive mammary neoplasia through an inflammatory and metabolic phenotype linked to mTOR activation. Cancer Res. 2013;73:4349–4361. doi: 10.1158/0008-5472.CAN-13-0322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Xu L., Han C., Lim K., Wu T. Cross-talk between Peroxisome Proliferator-Activated Receptor δ and Cytosolic Phospholipase A2α/Cyclooxygenase-2/Prostaglandin E2 Signaling Pathways in Human Hepatocellular Carcinoma Cells. Cancer Res. 2006;66:11859–11868. doi: 10.1158/0008-5472.CAN-06-1445. [DOI] [PubMed] [Google Scholar]
- 83.Peters J.M., Gonzalez F.J., Muller R. Establishing the Role of PPARbeta/delta in Carcinogenesis. Trends Endocrinol. Metab. 2015;26:595–607. doi: 10.1016/j.tem.2015.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Takayama O., Yamamoto H., Damdinsuren B., Sugita Y., Ngan C.Y., Xu X., Tsujino T., Takemasa I., Ikeda M., Sekimoto M., et al. Expression of PPAR[delta] in multistage carcinogenesis of the colorectum: Implications of malignant cancer morphology. Br. J. Cancer. 2006;95:889–895. doi: 10.1038/sj.bjc.6603343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Yoshinaga M., Taki K., Somada S., Sakiyama Y., Kubo N., Kaku T., Tsuruta S., Kusumoto T., Sakai H., Nakamura K., et al. The expression of both peroxisome proliferator-activated receptor delta and cyclooxygenase-2 in tissues is associated with poor prognosis in colorectal cancer patients. Dig. Dis. Sci. 2011;56:1194–1200. doi: 10.1007/s10620-010-1389-9. [DOI] [PubMed] [Google Scholar]
- 86.Zuo X., Xu W., Xu M., Tian R., Moussalli M.J., Mao F., Zheng X., Wang J., Morris J.S., Gagea M., et al. Metastasis regulation by PPARD expression in cancer cells. JCI Insight. 2017;2:e91419. doi: 10.1172/jci.insight.91419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Abdollahi A., Schwager C., Kleeff J., Esposito I., Domhan S., Peschke P., Hauser K., Hahnfeldt P., Hlatky L., Debus J., et al. Transcriptional network governing the angiogenic switch in human pancreatic cancer. Proc. Natl. Acad. Sci. USA. 2007;104:12890–12895. doi: 10.1073/pnas.0705505104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Pedchenko T.V., Gonzalez A.L., Wang D., DuBois R.N., Massion P.P. Peroxisome proliferator-activated receptor beta/delta expression and activation in lung cancer. Am. J. Respir. Cell Mol. Boil. 2008;39:689–696. doi: 10.1165/rcmb.2007-0426OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Harman F.S., Nicol C.J., Marin H.E., Ward J.M., Gonzalez F.J., Peters J.M. Peroxisome proliferator-activated receptor-delta attenuates colon carcinogenesis. Nat. Med. 2004;10:481–483. doi: 10.1038/nm1026. [DOI] [PubMed] [Google Scholar]
- 90.Wang D., Wang H., Guo Y., Ning W., Katkuri S., Wahli W., Desvergne B., Dey S.K., DuBois R.N. Crosstalk between peroxisome proliferator-activated receptor delta and VEGF stimulates cancer progression. Proc. Natl. Acad. Sci. USA. 2006;103:19069–19074. doi: 10.1073/pnas.0607948103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Zuo X., Peng Z., Moussalli M.J., Morris J.S., Broaddus R.R., Fischer S.M., Shureiqi I. Targeted Genetic Disruption of Peroxisome Proliferator-Activated Receptor-{delta} and Colonic Tumorigenesis. J. Natl. Cancer Inst. 2009;101:762–767. doi: 10.1093/jnci/djp078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He T.C., Chan T.A., Vogelstein B., Kinzler K.W. PPARdelta is an APC-regulated target of nonsteroidal anti-inflammatory drugs. Cell. 1999;99:335–345. doi: 10.1016/S0092-8674(00)81664-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Zuo X., Xu M., Yu J., Wu Y., Moussalli M.J., Manyam G.C., Lee S.I., Liang S., Gagea M., Morris J.S., et al. Potentiation of colon cancer susceptibility in mice by colonic epithelial PPAR-delta/beta overexpression. J. Natl. Cancer Inst. 2014;106:dju052. doi: 10.1093/jnci/dju052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Ghosh M., Ai Y., Narko K., Wang Z., Peters J.M., Hla T. PPARδ is pro-tumorigenic in a mouse model of COX-2-induced mammary cancer. Prostaglandins Other Lipid Mediat. 2009;88:97–100. doi: 10.1016/j.prostaglandins.2008.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Muller-Brusselbach S., Komhoff M., Rieck M., Meissner W., Kaddatz K., Adamkiewicz J., Keil B., Klose K.J., Moll R., Burdick A.D., et al. Deregulation of tumor angiogenesis and blockade of tumor growth in PPARbeta-deficient mice. EMBO J. 2007;26:3686–3698. doi: 10.1038/sj.emboj.7601803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Ham S.A., Yoo T., Hwang J.S., Kang E.S., Lee W.J., Paek K.S., Park C., Kim J.H., Do J.T., Lim D.S., et al. Ligand-activated PPARdelta modulates the migration and invasion of melanoma cells by regulating Snail expression. Am. J. Cancer Res. 2014;4:674–682. [PMC free article] [PubMed] [Google Scholar]
- 97.Her N.G., Jeong S.I., Cho K., Ha T.K., Han J., Ko K.P., Park S.K., Lee J.H., Lee M.G., Ryu B.K., et al. PPARdelta promotes oncogenic redirection of TGF-beta1 signaling through the activation of the ABCA1-Cav1 pathway. Cell Cycle. 2013;12:1521–1535. doi: 10.4161/cc.24636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Beyaz S., Mana M.D., Roper J., Kedrin D., Saadatpour A., Hong S.J., Bauer-Rowe K.E., Xifaras M.E., Akkad A., Arias E., et al. High-fat diet enhances stemness and tumorigenicity of intestinal progenitors. Nature. 2016;531:53–58. doi: 10.1038/nature17173. [DOI] [PMC free article] [PubMed] [Google Scholar]