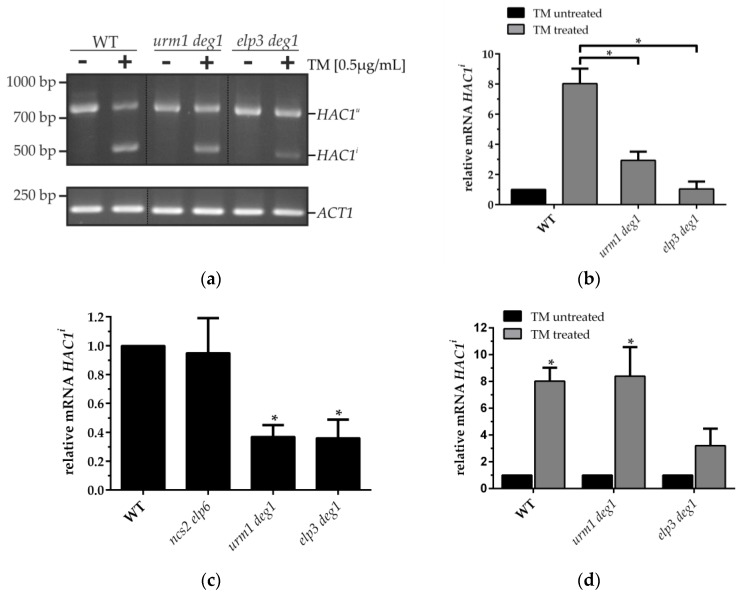
Figure 1.
Analysis of basal and tunicamycin (TM) induced HAC1 splicing in various transfer RNA (tRNA) modification mutants. (a) To measure the splicing of HAC1 mRNA in wildtype (WT), elp3 deg1 and urm1 deg1 cells RT-PCR was conducted as described [34]. In each sample, ACT1 mRNA was detected as a control. Yeast strains were cultivated in yeast peptone dextrose (YPD) until OD600 = 1.0 and harvested for RNA extraction. As a control, WT was additionally treated with TM (0.5 µg/mL) for 3 h (+). HAC1u represents the unspliced HAC1 mRNA and HAC1i the mature spliced HAC1 mRNA. (b–d) Quantification of the HAC1i mRNA level without (b), and with TM treatment (c,d) via qRT-PCR of the indicated strains. Induction of HAC1 splicing with TM was carried out as described in (a). mRNA levels were normalized to ACT1 using the ΔΔCt method [33]. The results obtained with TM treated yeast strains were standardized against the HAC1i mRNA level of the untreated wild-type (c) or the corresponding untreated strains (d), respectively. Quantitative PCR was performed with at least three biological triplicates per strain and condition and statistical significance was determined using a two-tailed t-test and indicated in the bar charts (* p < 0.05, b–d).