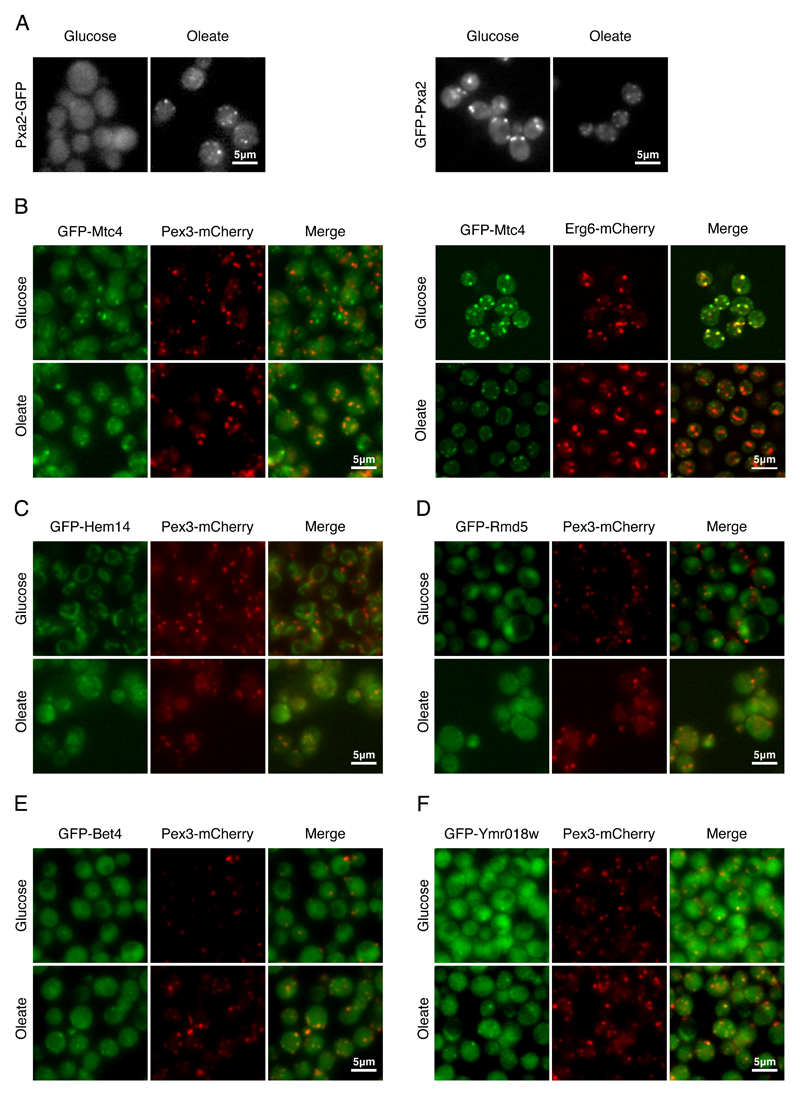
Figure 2. Identifying potential new peroxisomal proteins.
(A) When Pxa2-GFP was expressed under its native promoter it was observed in puncta only in oleate. However, when GFP-Pxa2 was expressed under a constitutive promoter, the protein was observed in puncta both in glucose and in oleate, implying that regulation is transcription-mediated. (B) Mtc4, a protein of unknown function, co-localized with a peroxisomal marker in oleate. In glucose Mtc4 co-localized with a lipid droplet marker, implying that Mtc4 localization is dependent on the carbon source of the cells. (C) Hem14, a mitochondrial enzyme that catalyzes the seventh step in the heme biosynthetic pathway, was the only mitochondrial protein that was partially co-localized with Pex3 in oleate. (D) Rmd5, a ubiquitin ligase, changed its localization from cytosol and nucleus in glucose to cytosol and puncta that co-localized with Pex3 in oleate. (E) Bet4, the α subunit of type II geranylgeranyltransferase, was co-localized with Pex3 during growth in glucose and oleate (F) Ymr018w, a protein whose function is unknown that shares similarity to the yeast Pex5, was co-localized with Pex3-mCherry both in glucose and in oleate. All images show a single focal plane.