Figure 4: Sequence dependence of fold and function.
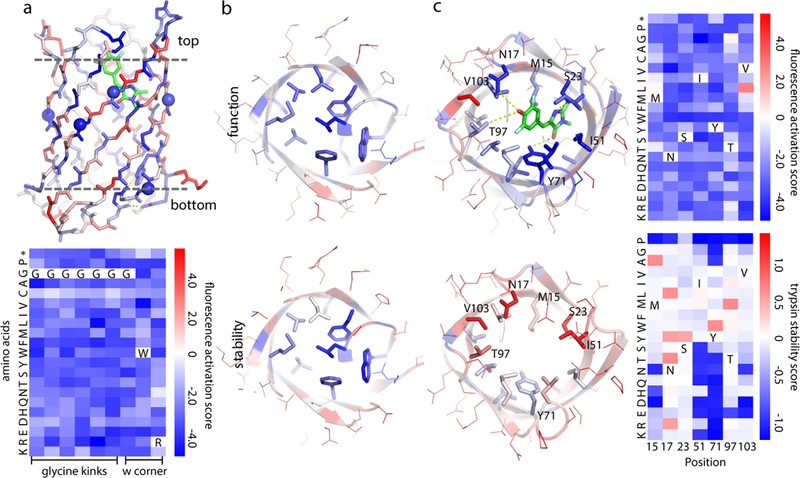
Each position was mutated one at a time to each of the other 19 amino acids, and the resulting library subjected to selection for fluorescence or stability to proteases. a, (Upper) b11L5F backbone model colored by relative fluorescence activation score at each position. Blue positions are strongly conserved during yeast selection; red positions are frequently substituted by other amino acids. Residues buried in the design model are much more conserved than solvent exposed residues. (Lower) All the mutations to the glycine kinks (spheres in the upper model) and tryptophan (W) corner considerably reduced fluorescence; the shape of the designed structure is critical for the designed function. b & c, Bottom (b) and top (c) comparisons of b11L5F side chains colored by relative fluorescence activation scores (upper row) and stability scores (lower row). In the bottom of the barrel, core residues were strongly conserved in both the function and stability selections (b); in the top barrel there is a clear function-stability trade-off with the key DFHBI interacting residues critical for function but far from optimal for stability (c, substitution patterns at these positions are shown on the right). Fluorescence activation and stability scores were derived from n=2 biologically independent experiments with >10-fold sequencing coverage. Standard deviation and confidence interval are provided in Extended Data Fig. 7.
