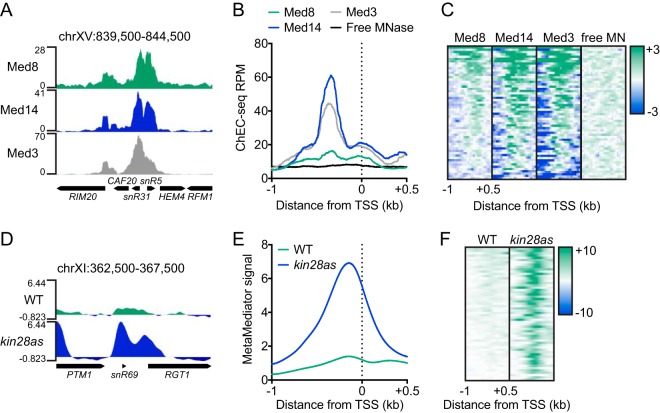
FIG 1.
Mediator associates with sn/snoRNA genes. (A) Genome browser view of the RPM-normalized Med8, Med14, and Med3 ChEC-seq signal at a representative sn/snoRNA-containing region of the yeast genome. (B) Average plot of the Med8, Med14, Med3, and free MNase ChEC-seq signal at 58 sn/snoRNA genes. (C) Heatmaps of the Med8, Med14, and Med3 ChEC-seq signal at 58 sn/snoRNA genes sorted in descending order by average Med8 signal. Heatmaps were log2 transformed with a center of 5 and are displayed with a contrast value of 3. (D) Genome browser view of the MetaMediator ChIP-chip signal in 1-NA-PP1-treated WT and kin28as cells at a representative sn/snoRNA-containing region of the yeast genome. (E) Average plot of the MetaMediator ChIP-chip signal in 1-NA-PP1-treated WT and kin28as cells at sn/snoRNA genes. (F) Heatmaps of the MetaMediator ChIP-chip signal at sn/snoRNA genes in WT and kin28as cells sorted in descending order by average WT signal. Heatmaps were not log2 transformed, as the raw data were provided in this form, and are displayed with a contrast value of 10. chr, chromosome.