Abstract
PISTILLATA (PI) homologs are crucial regulators of flower development in angiosperms. In this study, we isolated the MAwuPI homolog from Magnolia wufengensis, a basal angiosperm belonging to the Magnoliaceae. Molecular phylogenetic analysis suggested that MAwuPI was grouped into the PI/GLO lineages of B-class MADS-box gene with the distinctive PI motif. Further expression profiling analysis showed that MAwuPI was expressed in tepals and stamens but not in juvenile leaves and carpels, similar to the spatial expression pattern of AtPI in Arabidopsis. Interestingly, MAwuPI had higher expression level in inner-tepals than in outer-tepals, whereas the M. wufengensis flower is homochlamydeous. Moreover, ectopic expression of MAwuPI in Arabidopsis pi-1 mutant emerged filament-like structures but had no obvious petals, suggesting a partial phenotypic recovery of pi-1 mutant. The features of MAwuPI in the expression pattern and gene function improved our acknowledgment of B-class genes in M. wufengensis, and contributed to the clarification of M. wufengensis evolution status and relations with other sibling species in molecular perspective.
Keywords: PISTILLATA, Magnolia wufengensis, ABC model, flower development, ectopic expression
Introduction
Flowers, as the reproductive organ of plants, are the most complex structure of angiosperm (Endress, 2006). Compared to the reproductive structures of other plant groups, the flowers of angiosperm exhibit more variations in the number, morphology, size, color, and arrangement pattern of floral organs (Barrett, 2002). Thus, floral organ variation reflects angiosperm diversification. And the diversification of floral organ identity genes may have played a central role in the evolution of novel and different angiosperm morphologies (Irish, 2006).
The well-established ABCE model is widely accepted to explain the regular, sequential development of sepals, petals, stamens, and carpels in eudicot flowers (Bowman et al., 1991; Coen and Meyerowitz, 1991; Pelaz et al., 2000; Krizek and Fletcher, 2005). According to this model, the A-class genes determine the sepal and petal identity, the B-class genes specify petal and stamen identify, the C-class genes regulate stamen and carpel identity, and the E-class genes control the four whorl development. Moreover, the A-class and C-class genes function antagonistically. The combination of corresponding genes determines each floral organ identity. Although this model was built up on mutant studies in Arabidopsis and Antirrhinum, growing evidence showed that it may be suitable in all angiosperms (Theissen and Saedler, 2001; Kim et al., 2005; Melzer et al., 2010).
So far, most ABCE homeotic genes encode the MADS-box proteins, a huge transcriptional factor family (Becker and Theißen, 2003). MADS was named after its four major members, including MCM1, AGAMOUS, DEFICIENS, and SRF (Kim et al., 2004). Structural and molecular phylogenetic analysis indicated that plant MADS-box genes are belonging to MIKC-type MADS-box genes (Kaufmann et al., 2005), which contain four conserved domains, the MADS-, Intervening-, Keratin-like, and C-terminal (MIKC) domains (Becker and Theißen, 2003). The MADS domain, a highly conserved N-terminal domain, consists of about 58 amino acids (aa) and functions in dimerization by interaction with other transcriptional factors (Shore and Sharrocks, 1995). K domain, containing about 70 aa, is named because of the highly homologous primary structure with keratin. Three segments of α-helix (K1, K2, and K3) forms a coiled-coil secondary structure, involved in protein-protein interaction (Yang and Thomas, 2004). Between M domain and K domain lies the I domain (Krizek and Meyerowitz, 1996; Riechmann et al., 1996). The sequence and structure of the C-terminal region are highly variant, and play key roles in transcription factor activation, protein tetramerization, and functional specialization (Egea-Cortines et al., 1999; Honma and Goto, 2001; Lamb and Irish, 2003).
In Arabidopsis, APETALA1 (AP1) and AP2 belong to A-class genes, PISTILLATA (PI) and AP3 belong to B-class genes, AGAMOUS (AG) and AGAMOUS-LIKE (AGL) belong to C-class genes, and SEPALLATA1 (SEP1), SEP2, SEP3, and SEP4 belong to E-class genes (Ditta et al., 2004; Krizek and Fletcher, 2005). Mutation in AP2 gene results in the ectopic expression of C-class genes in outer two whorls, leading to sepal-to-carpel and petal-to-stamen homeotic transformation. On the contrary, in ag mutants, stamens converts into petals and another flower arises in place of the gynoecium (Bowman et al., 1991). Moreover, mutations in B-class genes convert the second whorl petal into a sepal structure and the third whorl stamen into a carpel structure (Goto and Meyerowitz, 1994). Recent researches further revealed that the ABC homeotic genes widely exist and are highly conserved among angiosperms (Becker and Theißen, 2003; Liu et al., 2010).
Magnolia wufengensis L.Y.Ma et L.R.Wang and M. wufengensis var. multitepala L.Y.Ma et L.R.Wang are newly identified species and variant, belonging to the Magnoliaceae, Magnolia (Ma et al., 2006). So far, M. wufengensis has only been found in mid-western Wufeng, Hubei Province in China, located in Wuling Mountain area, where many old plants originate and evolve. The limited distribution in wild and the local environmental disruption from human activities greatly restrict its regeneration in wild condition, and thus this species is critically endangered. However, the discovery of its wild population provides key information for identifying the center location of differentiation, distribution, and origination of modern Magnoliaceae family. Meanwhile, as a fine timber and landscape tree, as well as a precious medicinal tree, this species is also of great potential economic value. The flowers of M. wufengensis are highly different from core eudicots, like Arabidopsis. They have multiple morphologically identical petaloid tepals instead of distinct sepals and petals, and the numbers of the tepals vary from 9 to 24. In addition, the Manglietia plants as the relative early-diverging Magnoliaceae plants, have stable tepals, while the Michelia plants as the late-diverging Magnoliaceae plants, have largely varied tepals. And the Magnolia plants lies between the Manglietia and the Michelia plants during phylogenetic evolution. Thus, M. wufengensis L.Y.Ma et L.R.Wang and M. wufengensis var. multitepala L.Y.Ma et L.R.Wang provide ideal materials for the investigation on tepals development as well as the evolution and origin of the flowers in the Magnoliaceae plants.
B-class floral organ identity genes are key transcription factors controlling the development of petals and stamens (Kim et al., 2004). Previous studies have revealed that the diversity of flower organs in angiosperm is highly correlated with the variation of expression pattern and functional differentiation of B-class genes (Kim et al., 2004; Irish, 2006). Thus, phylogenetic and functional analysis of M. wufengensis PISTILLATA (MAwuPI) homolog may further reveal the mechanism of petal variation in M. wufengensis and clarify its evolution status in Magnolia plants.
Materials and Methods
Plant Material
Flower buds and juvenile leaves at different developmental stages were collected from M. wufengensis growing in the secondary natural forest at 1500 m elevation in Wufeng, southwest of Hubei Province. After sampled, flower buds were separated into outer-tepals, inner-tepals, stamens, and carpels (as shown in Supplementary Figure S1), and frozen immediately in liquid nitrogen.
The pi-1 mutant was obtained by EMS (ethyl methanesulfonate) mutagenesis in the genetic background Ler (Goto and Meyerowitz, 1994), which contains a point mutation causing Trp80 (TGG) to stop code (TGA). The pi-1 mutant and its corresponding wild-type Landsberg erecta were sown in soil and grown in controlled greenhouse at 22°C under 12 h light and 12 h dark cycles, with an irradiance of 120 μmol quanta m−2 s−1 and 70% relative humidity. Resistance selection and PCR-mediated genotyping were performed as Jing et al. (2015) previously described. The mature flowers of mutant and transgenic Arabidopsis plants were photographed by Leica165C stereomicroscope. For a better view, one of the petals has been removed before photographing.
Isolation of PI Homolog in M. wufengensis
Total RNA of M. wufengensis were extracted using EASYspin plant RNA Extraction Kit (Aidlab, Beijing) according to the manufacturer’s instructions. And for each sample, 1 μg RNA was used for cDNA synthesis with RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, Waltham, MA, United Ststes) as previously described (Jing et al., 2014, 2015). A 659 bp 3′ end of PI homolog from M. wufengensis was amplified from the cDNA through 3′ RACE-specific primer GSPPI and 3′ RACE Outer Primer by the 3′-full RACE Core Set Ver.2.0 kit (TaKaRa, Japan), and a 910 bp 5′ end was amplified through 5′ RACE-specific primer PIGSP1 and PIGSP2 by 5′-full RACE Kit (TaKaRa, Japan). After assembly, the integrity of the cDNA sequences of the PI homolog from M. wufengensis were then verified using the forward primer MawuPIF and the reverse primer MawuPIR. PCR was performed with a 5 min at 94°C denaturation step, followed by 30 cycles of 45 s at 94°C, 45 s annealing at 56°C, 45 s extending at 72°C, with a final extension period of 10 min at 72°C. These PCR products were subcloned into pMD18-T Vector (TaKaRa, Japan) and sequenced. The primers used for isolation of PI homolog in M. wufengensis were listed in Supplementary Table S1.
Sequence Alignments and Phylogenetic Analysis
Deduced amino acid sequences of MawuPI were blasted on the GenBank database. The molecular weight (MW) and theoretical isoelectric point (pI) of the coded protein were analyzed using the online ProtParam tool1 (Gasteiger et al., 2005). During the BLAST searches, multiple B class proteins from various angiosperm lineages were selected for alignment. Full-length amino acid sequences of these genes were aligned with a ClustalW program under default settings (Thompson et al., 1994) and phylogenetic trees were constructed by MEGA 5.0 software (Tamura et al., 2011) using the Neighbor-Joining (NJ) and Maximum Likelihood (ML) method with bootstrap setting as 1000 replicates. The accession numbers of the sequence used for phylogenetic analysis were listed in Supplementary Table S2.
Semi-Quantitative and Quantitative RT-PCR
To analyze the expression level of MawuPI in different organs, total RNA was extracted from leaf, outer-tepals, inner-tepals, stamens and carpels of M. wufengensis as described above. Semi-quantitative PCR was performed as follows: 5 min at 94°C, 25 cycles of 30 s at 94°C, 30 s at 56°C, 30 s at 72°C, followed by 10 min at 72°C. PCR products was analyzed by electrophoresis in a 1% agarose gel and photographed under UV light. Then, quantitative RT-PCR was performed on IQ5 Multicolor Real-time PCR Detection System (Bio-Rad) to analyze the expression level of MawuPI in developmental stages. The reaction mixture was cycled as follows: 95°C for 3 min, followed by 40 cycles of 95°C for 10 s, 55°C for 20 s, 72°C for 20 s; the temperature changed by 0.5°C/s to 95°C for the melt curve. The MAwuACTIN gene was chosen as the internal control as previously reported (Wu et al., 2012). All experiments were performed with at least three biological replicates and three technical replicates. The gene specific primers used for semi-quantitative RT-PCR and quantitative RT-PCR were designed according to the isolated cDNA sequence. And the primer sequences were listed in Supplementary Table S1.
Vector Construction and Arabidopsis Transformation
Full-length MAwuPI cDNA were amplified and cloned into a binary vector pBI121 (BD Biosciences Clontech) using XbaI and SmaI restriction enzymes. The 35S::MAwuPI construction was transformed into heterozygous pi-1 plant using the Agrobacterium-mediated floral-dip method (Clough and Bent, 1998). The transgenic Arabidopsis seeds were selected on a solid half-strength MS medium containing 50 μg/ml kanamycin at 4°C for 2 days, which were then transferred to the greenhouse for additional 10 days. Subsequently, the green seedlings were selected as positive seedlings and transplanted in soil. Then, genomic PCR and qRT-PCR were carried out to further confirm the 35S::MAwuPI transgenic lines as previously described (Jing et al., 2015). Meanwhile, genotyping of wild-type, heterozygote and homozygous pi-1 transformants was performed using a dCAPS marker (Neff et al., 2002; Jing et al., 2015). The primers used for vector construction and genotyping were listed in Supplementary Table S1.
Results
Cloning and Sequence Analysis of MAwuPI
To obtain the sequence of MAwuPI, homology-based cloning and RACE techniques were carried out to identify the full-length cDNA sequence. According to the assemble sequence, we further designed specific primers to amplify and verify the sequence. And we found that the cDNA of MAwuPI contains 1384 bp, including 573 bp of 5′-UTR, 172 bp of 3′-UTR, and 639 bp of ORF encoding for 212 amino acids. The molecular weight (MW) and isoelectric points (PI) of MAwuPI protein is 25 kD and 9.51, respectively. The sequence was deposited in GenBank under accession number JN315682.
Further protein sequence comparisons were performed with seven PI/GLO proteins as well as a DEF/AP3 protein (AtAP3) as a contrast. Meanwhile, we have combined the phylogenetic analysis with a schematic expression in whorls of the selected genes (Figure 1). Our results showed that this protein contains a highly conserved MADS domain (1–59aa), a less conserved K domain (88–170aa), a least conserved I region (60–87aa) between M and K domain, and a highly conserved PI motif lying in C-terminal region (171–212aa) (Figure 1), which is essential for the formation of heterologous tetramer and functional specialization (Lamb and Irish, 2003; Kaufmann et al., 2005; Liu et al., 2010).
FIGURE 1.
Comparison of deduced amino acid sequences encoded by MAwuPI and related members of the PI/GLO lineage. Dots indicate gaps inserted for alignment optimization. Sequence comparison of MAwuPI and the PI/GLO-related MADS domain proteins. The PI motif in the C-terminal region is boxed. The motif is highly conserved for PI/GLO lineage proteins with a DEF/AP3 protein (AtAP3) as a contrast. Meanwhile, a schematic expression of the selected genes in the four whorls of the flowers is also combined.
Phylogenetic Analysis
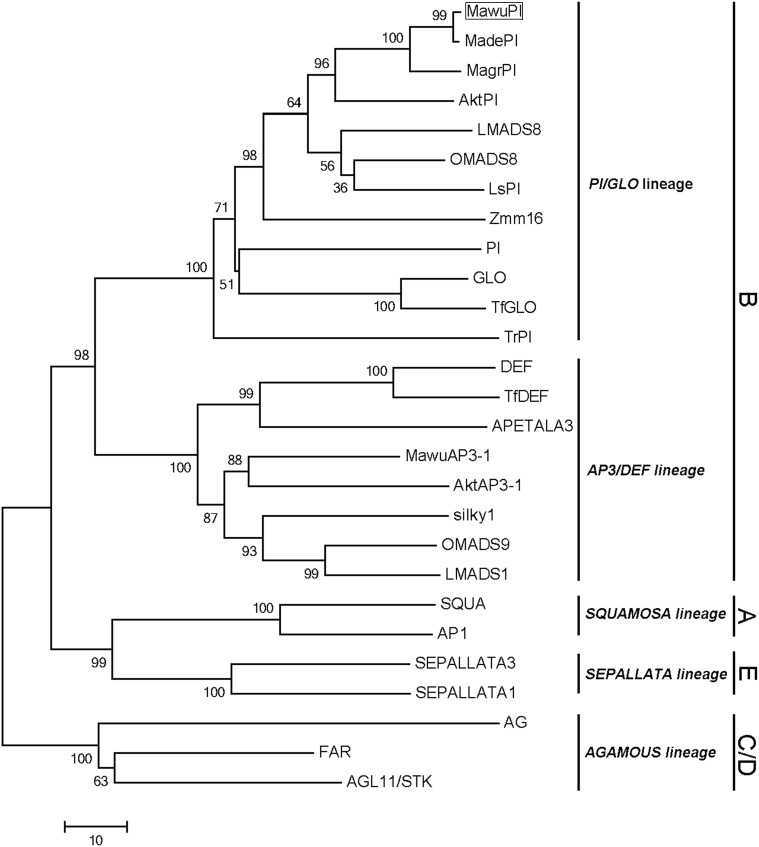
To reveal the evolution of MAwuPI, the cloned MAwuPI sequence was then blasted with twenty B-class genes from other angiosperms (Supplementary Table S2). Meanwhile, two A-class genes, two B-class genes, one D-class gene, and two E-class genes were chosen as outgroup (Supplementary Table S2). Phylogenetic analysis suggested that MAwuPI is grouped into the PI/GLO lineages of B-class genes, together with other eleven PI/GLO lineage genes (Figure 2).
FIGURE 2.
Phylogenetic analysis of PI/GLO lineage. The MAwuPI protein sequence is blasted with twenty B-class genes from other angiosperms, with two A-class genes, two C-class genes, one D-class gene and two E-class genes as outgroup. The MAwuPI is boxed.
Expression Profiling of MAwuPI in M. wufengensis
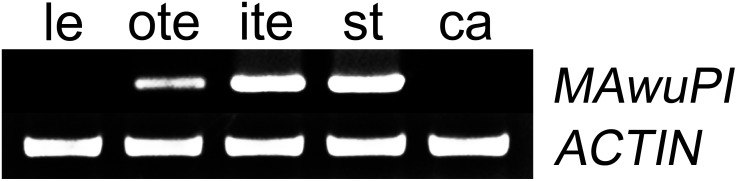
Since A-, B-, and C-class floral organ identity genes are expression in distinct floral whorls, we firstly analyzed the expression pattern of MAwuPI in different floral organs by semi-quantitative RT-PCR. As shown in Figure 3, MAwuPI was expressed in tepals and stamens, but not in juvenile leaves and carpels. This is similar to the spatial expression pattern of AtPI in Arabidopsis (Goto and Meyerowitz, 1994).
FIGURE 3.
Semi-quantitative PCR analysis of the MAwuPI expression pattern in Magnolia wufengensis. For this assay, flower buds, and juvenile leaves were collected from M. wufengensis growing in the secondary natural forest. The flower buds were separated into outer-tepals, inner-tepals, stamens, and carpels, and frozen immediately in liquid nitrogen. Three biological replicates were performed for each sample. le, leaf; te, tepal; st, stamen; ca, carple; actin as internal control.
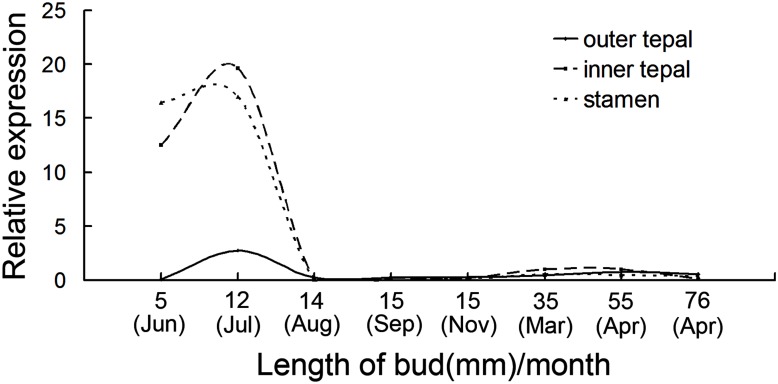
Next, quantitative RT-PCR was performed to monitor the dynamic changes of MAwuPI expression in out-tepals, inner-tepals, and stamens during different developmental stages. In June and July, when the primordium of inner-tepals and stamens initiated and grown fast, the expression of MAwuPI in inner-tepals and stamens elevated quickly and reached the peak in July when the flower buds were about 12 mm in length; in July and August, when the temperature continued to rise in southwest of Hubei province and the development of flower buds consequently slowed down, the expression of MAwuPI in inner-tepals and stamens decreased quickly (Figure 4). Although the expression of MAwuPI in out-tepals was comparatively lower, the variation trend matched those in inner-tepals and stamens (Figure 4). Additionally, from August onward, when the flower buds entered dormancy, the expression of MAwuPI in three whorls maintained in an extremely low level. Until March the following year, before flowering, its expression increased a little, but still largely lower than the peak (Figure 4).
FIGURE 4.
Expression pattern of MAwuPI at different developmental stages. For this assay, plant materials were collected from M. wufengensis growing in the secondary natural forest at different time-point, and frozen immediately in liquid nitrogen. Three biological replicates were performed at each time-point, and at least three technical replicates were performed for each data. Gene expression level was shown as fold changes in comparison to MAwuactin.
Ectopic Expression of MAwuPI Partially Rescues the Floral Deficiency of pi-1 Mutant
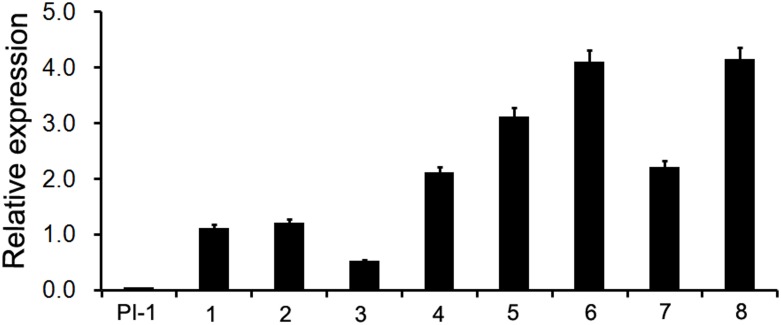
To further analyze the function of MAwuPI, complementation assay was performed by ectopic expression of MAwuPI into Arabidopsis pi-1 mutant. The homozygous pi-1 mutant (Figure 5a) is a strong allele expressing a truncated protein product of 79 aa, resulting in petals altered to sepals and stamens altered to carpels, while the heterozygote pi-1 mutant (Figure 5c) exhibited a wild-type phenotype (Figure 5b; Goto and Meyerowitz, 1994). The transgenic plants were selected by kanamycin plates and PCR-mediated genotyping. A total of eight independent lines were obtained and assayed to monitor the expression levels of MAwuPI by qRT-PCR (Figure 6). Morphological observations showed that compared to pi-1 mutant, five transgenic lines (4#, 5#, 6#, 7#, and 8#) ectopically expressing MAwuPI (Figures 5d–h) emerged filament-like structures in the third whorl of floral organ. This observation indicated that constitutive expression of MAwuPI can rescue the floral deficiency of pi-1 mutant to different degrees. However, the second whorl was not obviously recovered (Figure 5), which implied that MAwuPI complemented the pi-1 mutant only partially. Thus, these results suggested that MAwuPI plays a key role in stamen development.
FIGURE 5.
The mature flowers of transgenic plants ectopically expressing MAwuPI in Arabidopsis pi-1 mutant. (a) The flower of pi-1 homozygous Arabidopsis thaliana. (b) The flower of wild-type Arabidopsis thaliana. (c) The flower of pi-1 heterozygous Arabidopsis thaliana. For a better view, one of the petals has been removed. (d–h) 35S::MAwuPI transgenic Arabidopsis in pi-1 mutant background (line 4#, 5#, 6#, 7#, and 8#, respectively).
FIGURE 6.
Quantitative Real-time PCR analysis of the MAwuPI transcript levels in complementary lines. At least three biological replicates and three technical replicates were performed for each data. Gene expression level was shown as fold changes, and the gene expression level in wild-type Arabidopsis plants was set as 1. Error bars represent standard error of the means of three biological repeats. Atactin is used as internal control. PI-1, pi-1 mutant; 1–8, different 35S::MAwuPI transgenic lines in Arabidopsis pi-1 mutant background.
Discussion
In most core eudicots, PISTILLATA homologous genes, encoding the B-class transcription factors, are key regulators of petal and stamen development. In this study, we isolated and analyzed the expression patterns of MAwuPI during floral organ development and phylogenetic evolution between its closely related species. Meanwhile, we carried out functional complement experiments in Arabidopsis to further confirm its functions in specifying floral organ identities.
Though homology-based cloning and RACE techniques, we obtained the full-length cDNA sequence of MAwuPI. Molecular phylogenetic analysis suggested that MAwuPI belongs to the PI/GLO lineages of B-class MADS-box gene, since it contains the distinctive PI motif (Kim et al., 2004). Also, MAwuPI protein contains a highly conserved MADS domain, a less-conserved K domain, a least conserved I domain and a highly variant C-terminal domain. Thus, according to the sequence, MAwuPI is a typical B-class MADS-box gene.
The floral organ of a typical core eudicot composes of four whorls outside-to-inside, including sepals, petals, stamens, and carpels. In wild-type Arabidopsis, the expression of PI initiates after the formation of sepal primordium, which is firstly detected in the inner three whorls and later restricted in the second and third whorls of the floral organ. The silenced expression of PI in the fourth whorl during floral organ development might be due to the dependence between the expression of PI and AP3, which together confer B-class gene function (Jack et al., 1992; Goto and Meyerowitz, 1994; Krizek and Meyerowitz, 1996). Here, we found that MAwuPI was expressed in petaloid tepals and stamens (Figure 3), in accordance with the feature of B-class genes. Interestingly, the expression level of MAwuPI was dramatically higher in inner-tepals than in outer-tepals although the M. wufengensis flower is homochlamydeous. This expression pattern in M. wufengensis is different from that in Arabidopsis as well as other species (Figure 1). For instance, LMADS8, the PI homologous gene from Lilium longiflorum, was only expressed in stamens at early stage of flower bud differentiation, and then diffused to outer- and inner-tepals with equal expression level in these two whorls (Chen et al., 2012). In Lacandonia schismatica, the transcriptional activity of LS-PI was restricted in stamens and carpels but not in tepals, together with LS-AP3 to regulate stamen development (Álvarez-Buylla et al., 2010). In Oncidium Gower Ramsey, the PI homologous gene OMADS8 was expressed in vegetative organs and all whorls of floral organ, and regulated tepals differentiation in coordination with other MADS-box genes (Chang et al., 2010; Mao et al., 2015). In Taihangia rupestris, TrPI was expressed in both vegetative and reproductive organ, with top expression level in petals and stamens (Lu et al., 2010). In Sesuvium portulacastrum, SpPI was only expressed in stamens but not in tepals, and regulated the formation of stamen rather than tepals during floral organ development (Brockington et al., 2012). In basal eudicot Eschscholzia californica, EScaGLO was expressed in petals and stamens, modulating their development (Lange et al., 2013). Taken together, in most angiosperms, the PI homologs participate in regulating petals (petaloid tepals) and stamen development, while in a few cases (like Lacandonia schismatica) they are only involved in stamen formation. However, the expression of PI homologs is not limited in the tissues where they functions, indicating that the expression pattern and biological function of PI homologs undergo multiply evolutions during the origin and evolution of angiosperms.
In M. wufengensis, MawuPI was mainly expressed in inner-tepals and stamens (Figure 3). Functional complementation assay showed that ectopic expression of MawuPI in Arabidopsis pi-1 mutant emerged filament-like structures in the third whorl of floral organ, whereas petals were still defect (Figure 5). These observations indicated that MawuPI may mainly regulate stamens formation, but may not contribute to petal development or else may require other regulator(s). The PI homologs from gymnosperms have also been reported the similar phenotypes (Winter et al., 2002a; Wang et al., 2010), which supported the functional conservation of PI homologs during evolution. In addition, LMADS8 was firstly expressed in stamens and then diffused to outer- and inner-tepals in monocotyledon Lilium longiflorum. And ectopic expression of LMADS8 could significantly recover stamens defect and partially recover petals defect of pi-1 (Chen et al., 2012). Similar results were also observed in Lacandonia schismatica of the Triuridaceae (Álvarez-Buylla et al., 2010). Besides, in Taihangia rupestris of the Rosaceae, ectopic expression of TrPI could restore the phenotypic defects of both petals and stamens in Arabidopsis pi-1 mutant (Lu et al., 2010). Based on these data, we may conclude that during the evolution from gymnosperms to angiosperms, PI homologs were initially involved in stamens development, and then functioned in tepals development with the expression zone expanded.
Functional analyses have suggested that the involvement of PI homologs in controlling stamens development is quite conserved between species, but their roles in regulating petals development differ. In Sesuvium portulacastrum of the Aizoaceae, SpPI was only expressed in and regulated stamens (Brockington et al., 2012). In Lacandonia schismatica of the Triuridaceae, LS-PI was expressed in stamens and carpels but LS-AP3 was only expressed in stamens, and the B-class genes mainly involved in specifying stamen identity (Álvarez-Buylla et al., 2010). Moreover, in most eudicots, PI homologs exert their function in modulating stamens and petals development, but their expression patterns in these species are largely varied (Lu et al., 2010; Sasaki et al., 2010; Lange et al., 2013; Fang et al., 2015). In general, their expression regions are much larger than where they exert their function. In other words, during evolution, floral organ identity genes expand their expression to new tissues, interact with local genes, and thereafter participate in organogenesis of these regions. In the Orchidaceae plants, as the group with most morphological diversity in monocotyledon, the tepals differentiation is highly correlated with the functions of B-class genes, which further supports this hypothesis (Chang et al., 2010; Mondragón-Palomino and Theissen, 2011). Interestingly, MawuPI has a higher expression level in stamens and inner-tepals than in outer-tepals, whereas the inner- and outer-tepals have no obvious morphological difference in M. wufengensis. However, heterodimeric complexes of DEF/AP3 and GLO/PI proteins are required for the development of tepals and stamens (Winter et al., 2002b). Thus, overexpressing only one of the two components might not be sufficient to recapitulate the function of MAwuPI in a wild-type context. On the other hand, although 35S promoter-drived complementation assay is widely used, regulatory elements necessary for correct MAwuPI expression are missing, which might affect the functions of MAwuPI. Thus, the mechanisms underlying MAwuPI-mediated tepals development remain to be further investigated.
Taken together, MawuPI was isolated and its functions in specifying floral organ identities were further characterized in this study. These data improved our knowledge of B-class genes in M. wufengensis, and contributed to the clarification of M. wufengensis evolution status and relations with other sibling species in molecular perspective.
Author Contributions
FC conceived and directed this study. WL, XS, HL, and YW performed the experiments. WL and FC wrote the manuscript. WL, XS, ZH, and DZ analyzed the data and revised the manuscript. All authors approved the manuscript and the version to be published, and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Yongfu Fu for providing us the Landsberg erecta wild-type and pi-1 mutant, as well as Agrobacterium tumefacieus strain GV1301-90. We also thank Prof. Hai Lu for providing us the binary vector pBI121.
Funding. This study was supported by the Natural Science Foundation of China (Grant No. 31170625).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2018.01743/full#supplementary-material
Morphological observation of outer-tepals and inner-tepals in the flower of M. wufengensis.
Primer sequences used in this study.
Names and accession numbers of protein sequences used for phylogenetic analysis.
References
- Álvarez-Buylla E. R., Ambrose B. A., Flores-Sandoval E., Englund M., Garay-Arroyo A., García-Ponce B., et al. (2010). B-function expression in the flower center underlies the homeotic phenotype of Lacandonia schismatica (Triuridaceae). Plant Cell 22 3543–3559. 10.1105/tpc.109.069153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett C. H. (2002). The evolution of plant sexual diversity. Nature 3 274–284. [DOI] [PubMed] [Google Scholar]
- Becker A., Theißen G. (2003). The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol. Phylogenet. Evol. 29 464–489. 10.1016/S1055-7903(03)00207-0 [DOI] [PubMed] [Google Scholar]
- Bowman J. L., Smyth D. R., Meyerowitz E. M. (1991). Genetic interactions among floral homeotic genes of arabidopsis. Development 112 1–20. [DOI] [PubMed] [Google Scholar]
- Brockington S. F., Rudall P. J., Frohlich M. W., Oppenheimer D. G., Soltis P. S., Soltis D. E. (2012). ‘Living stones’ reveal alternative petal identity programs within the core eudicots. Plant J. 69 193–203. 10.1111/j.1365-313X.2011.04797.x [DOI] [PubMed] [Google Scholar]
- Chang Y. Y., Kao N. H., Li J. Y., Hsu W. H., Liang Y. L., Wu J. W., et al. (2010). Characterization of the possible roles for B class MADS box genes in regulation of perianth formation in orchid. Plant Physiol. 152 837–853. 10.1104/pp.109.147116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. K., Hsieh W. P., Yang C. H. (2012). Functional analysis reveals the possible role of the C-terminal sequences and PI motif in the function of lily (Lilium longiflorum) PISTILLATA (PI) orthologues. J. Exp. Bot. 63 941–961. 10.1093/jxb/err323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough S. J., Bent A. F. (1998). Floral dip: a simplified method for agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16 735–743. 10.1046/j.1365-313x.1998.00343.x [DOI] [PubMed] [Google Scholar]
- Coen E. S., Meyerowitz E. M. (1991). The war of the whorls: genetic interactions controlling flower development. Nature 353 31–37. 10.1038/353031a0 [DOI] [PubMed] [Google Scholar]
- Ditta G., Pinyopich A., Robles P., Pelaz S., Yanofsky M. F. (2004). The SEP4 gene of Arabidopsis thaliana functions in floral organ and meristem identity. Curr. Biol. 14 1935–1940. 10.1016/j.cub.2004.10.028 [DOI] [PubMed] [Google Scholar]
- Egea-Cortines M., Saedler H., Sommer H. (1999). Ternary complex formation between the MADS-box proteins squamosa, deficiens and globosa is involved in the control of floral architecture in Antirrhinum majus. EMBO J. 18 5370–5379. 10.1093/emboj/18.19.5370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Endress P. K. (2006). Angiosperm floral evolution: morphological developmental framework. Adv. Bot. Res. 44 1–61. 10.1016/S0065-2296(06)44001-5 [DOI] [Google Scholar]
- Fang Z. W., Li X. P., Li X. F., Liu Z. X. (2015). FaesPI, a Fagopyrum esculentum PISTILLATA ortholog, is involved only in stamen development. J. Plant Biol. 58 102–109. 10.1007/s12374-014-0390-9 [DOI] [Google Scholar]
- Gasteiger E., Hoogland C., Gattiker A., Duvaud S., Wilkins M. R., Appel R. D., et al. (2005). “The proteomics protocols handbook - chapter 52: protein identification and analysis tools on the ExPASy server,” in The Proteomics Protocols Handbook, ed. Walker J. M. (New York, NY: Humana press; ) 571–607. [Google Scholar]
- Goto K., Meyerowitz E. M. (1994). Function and regulation of the arabidopsis floral homeotic gene PISTILLATA. Genes Dev. 8 1548–1560. 10.1101/gad.8.13.1548 [DOI] [PubMed] [Google Scholar]
- Honma T., Goto K. (2001). Complexes of MADS-box proteins are sufficient to convert leaves into floral organs. Nature 409 525–529. 10.1038/35054083 [DOI] [PubMed] [Google Scholar]
- Irish V. F. (2006). Duplication, diversification, and comparative genetics of angiosperm MADS-box genes. Adv. Bot. Res. 44 130–151. 10.1016/S0065-2296(06)44003-9 [DOI] [Google Scholar]
- Jack T., Brockman L. L., Meyerowitz E. M. (1992). The homeotic gene APETALA3 of Arabidopsis thaliana encodes a MADS box and is expressed in petals and stamens. Cell 68 683–697. 10.1016/0092-8674(92)90144-2 [DOI] [PubMed] [Google Scholar]
- Jing D., Liu Z., Zhang B., Ma J., Han Y., Chen F. (2014). Two ancestral APETALA3 homologs from the basal angiosperm Magnolia wufengensis (Magnoliaceae) can affect flower development of arabidopsis. Gene 537 100–107. 10.1016/j.gene.2013.11.076 [DOI] [PubMed] [Google Scholar]
- Jing D., Xia Y., Chen F., Wang Z., Zhang S., Wang J. (2015). Ectopic expression of a Catalpa bungei (Bignoniaceae) PISTILLATA homologue rescues the petal and stamen identities in arabidopsis pi-1 mutant. Plant Sci. 231 40–51. 10.1016/j.plantsci.2014.11.004 [DOI] [PubMed] [Google Scholar]
- Kaufmann K., Melzer R., Theissen G. (2005). MIKC-type MADS-domain proteins: structural modularity, protein interactions and network evolution in land plants. Gene 347 183–198. 10.1016/j.gene.2004.12.014 [DOI] [PubMed] [Google Scholar]
- Kim S., Koh J., Yoo M. J., Kong H., Hu Y., Ma H., et al. (2005). Expression of floral MADS-box genes in basal angiosperms: implications for the evolution of floral regulators. Plant J. 43 724–744. 10.1111/j.1365-313X.2005.02487.x [DOI] [PubMed] [Google Scholar]
- Kim S., Yoo M. J., Albert V. A., Farris J. S., Soltis P. S., Soltis D. E. (2004). Phylogeny and diversification of B-function MADS-box genes in angiosperms: evolutionary and functional implications of a 260-million-year-old duplication. Am. J. Bot. 91 2102–2118. 10.3732/ajb.91.12.2102 [DOI] [PubMed] [Google Scholar]
- Krizek B. A., Fletcher J. C. (2005). Molecular mechanisms of flower development: an armchair guide. Nature Rev. Genet. 6 688–698. 10.1038/nrg1675 [DOI] [PubMed] [Google Scholar]
- Krizek B. A., Meyerowitz E. M. (1996). Mapping the protein regions responsible for the functional specificities of the arabidopsis MADS domain organ-identity proteins. Proc. Natl. Acad. Sci. U.S.A. 93 4063–4070. 10.1073/pnas.93.9.4063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb R. S., Irish V. F. (2003). Functional divergence within the APETALA3/PISTILLATA floral homeotic gene lineages. Proc. Natl. Acad. Sci. U.S.A. 100 6558–6563. 10.1073/pnas.0631708100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange M., Orashakova S., Lange S., Melzer R., Theißen G., Smyth D. R., et al. (2013). The seirena B class floral homeotic mutant of California Poppy (Eschscholzia californica) reveals a function of the enigmatic PI motif in the formation of specific multimeric MADS domain protein complexes. Plant Cell 25 438–453. 10.1105/tpc.112.105809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C., Zhang J., Zhang N., Shan H., Su K., Zhang J., et al. (2010). Interactions among proteins of floral MADS-box genes in basal eudicots: implications for evolution of the regulatory network for flower development. Mol. Biol. Evol. 27 1598–1611. 10.1093/molbev/msq044 [DOI] [PubMed] [Google Scholar]
- Lu S., Fan Y., Liu L., Liu S., Zhang W., Meng Z. (2010). Ectopic expression of TrPI, a Taihangia rupestris (Rosaceae) PI ortholog, causes modifications of vegetative architecture in arabidopsis. J. Plant Physiol. 167 1613–1621. 10.1016/j.jplph.2010.06.028 [DOI] [PubMed] [Google Scholar]
- Ma L. Y., Wand L. R., He S. C., Liu X., Wang X. Q. (2006). A new species of Magnolia (Magnoliaceae) from Hubei, China. Bull. Bot. Res. 26 5–8. [Google Scholar]
- Mao W. T., Hsu H. F., Hsu W. H., Li J. Y., Lee Y. I., Yang C. H. (2015). The C-Terminal sequence and PI motif of the orchid (Oncidium Gower Ramsey) PISTILLATA (PI) ortholog determine its ability to bind AP3 orthologs and enter the nucleus to regulate downstream genes controlling petal and stamen formation. Plant Cell Physiol. 56 2079–2099. [DOI] [PubMed] [Google Scholar]
- Melzer R., Wang Y. Q., Theissen G. (2010). The naked and the dead: the ABCs of gymnosperm reproduction and the origin of the angiosperm flower. Semin. Cell Dev. Biol. 21 118–128. 10.1016/j.semcdb.2009.11.015 [DOI] [PubMed] [Google Scholar]
- Mondragón-Palomino M., Theissen G. (2011). Conserved differential expression of paralogous deficiens and globosa-like MADS-box genes in the flowers of orchidaceae: refining the ‘orchid code’. Plant J. 66 1008–1019. 10.1111/j.1365-313X.2011.04560.x [DOI] [PubMed] [Google Scholar]
- Neff M. M., Turk E., Kalishman M. (2002). Web-based primer design for single nucleotide polymorphism analysis. Trends Genet. 18 613–615. 10.1016/S0168-9525(02)02820-2 [DOI] [PubMed] [Google Scholar]
- Pelaz S., Ditta G. S., Baumann E., Wisman E., Yanofsky M. F. (2000). B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405 200–203. 10.1038/35012103 [DOI] [PubMed] [Google Scholar]
- Riechmann J. L., Krizek B. A., Meyerowitz E. M. (1996). Dimerization specificity of arabidopsis MADS domain homeotic proteins APETALA1, APETALA3, PISTILLATA, and AGAMOUS. Proc. Natl. Acad. Sci. U.S.A. 93 4793–4798. 10.1073/pnas.93.10.4793 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Aida R., Yamaguchi H., Shikata M., Niki T., Nishijima T., et al. (2010). Functional divergence within class B MADS-box genes TfGLO and TfDEF in Torenia fournieri Lind. Mol. Genet. Genomics 284 399–414. 10.1007/s00438-010-0574-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shore P., Sharrocks A. D. (1995). The MADS-box family of transcription factors. Eur. J. Biochem. 229 1–13. 10.1111/j.1432-1033.1995.tb20430.x [DOI] [PubMed] [Google Scholar]
- Tamura K., Peterson D., Peterson N., Stecher G., Nei M., Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28 2731–2739. 10.1093/molbev/msr121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theissen G., Saedler H. (2001). Floral quartets. Nature 409 469–471. 10.1038/35054172 [DOI] [PubMed] [Google Scholar]
- Thompson J. D., Higgins D. G., Gibson T. J. (1994). CLUSTALW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, positions-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22 4673–4680. 10.1093/nar/22.22.4673 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y. Q., Melzer R., Theissen G. (2010). Molecular interactions of orthologues of floral homeotic proteins from the gymnosperm Gnetum gnemon provide a clue to the evolutionary origin of ‘floral quartets’. Plant J. 64 177–190. 10.1111/j.1365-313X.2010.04325.x [DOI] [PubMed] [Google Scholar]
- Winter K. U., Saedler H., Theissen G. (2002a). On the origin of class B floral homeotic genes: functional substitution and dominant inhibition in Arabidopsis by expression of an orthologue from the gymnosperm Gnetum. Plant J. 31 457–475. 10.1046/j.1365-313X.2002.01375.x [DOI] [PubMed] [Google Scholar]
- Winter K. U, Weiser C., Kaufmann K., Bohne A., Kirchner C., Kanno A., et al. (2002b). Evolution of class B floral homeotic proteins: obligate heterodimerization originated from homodimerization. Mol. Biol. Evol. 19 587–596. 10.1093/oxfordjournals.molbev.a004118 [DOI] [PubMed] [Google Scholar]
- Wu W., Chen F., Jing D., Liu Z., Ma L. (2012). Isolation and characterization of an agamous-like gene from Magnolia wufengensis (Magnoliaceae). Plant Mol. Biol. Rep. 30 690–698. 10.1007/s11105-011-0385-3 [DOI] [Google Scholar]
- Yang Y., Thomas J. (2004). Defining subdomains of the K domain important for protein-protein interactions of plant MADS proteins. Plant Mol. Biol. 55 45–59. 10.1007/s11103-004-0416-7 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Morphological observation of outer-tepals and inner-tepals in the flower of M. wufengensis.
Primer sequences used in this study.
Names and accession numbers of protein sequences used for phylogenetic analysis.