Figure 4:
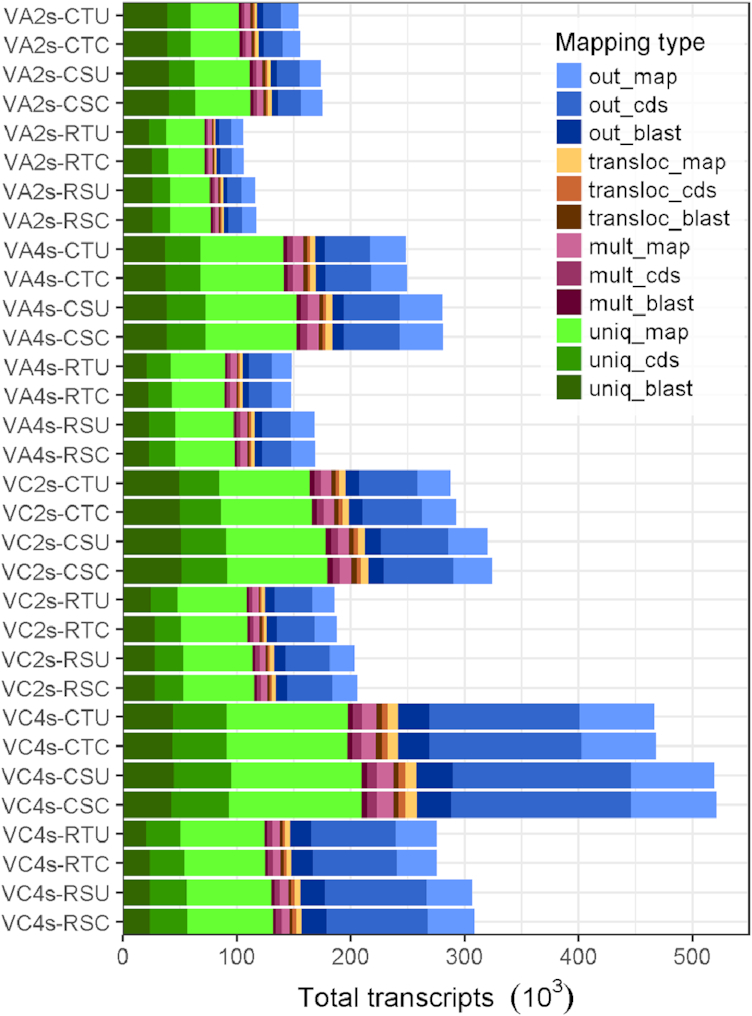
Mapping of de novo assembly transcriptomes to V.corymbosum reference genome and annotation of transcripts. Transcripts mapped uniquely to the genome (uniq), to multiple locations (mult), with translocations (transloc), or did not map (out). Annotation from prediction of coding sequences (cds) using homology results from blast is divided as “No Functional Annotation” (map), “CDS Only” (cds), and “CDS with Blast Hit” (blast). Transcriptomes for V. arboreum (VA) or V. corymbosum (VC) produced from two (2s) or four (4s) samples were clustered with either CD-HIT (C) or RapClust (R). The last two letters indicate trimming with Trimmomatic (T) or Skewer (S), and use (C) or not (U) of error correction of RNA-seq reads.
