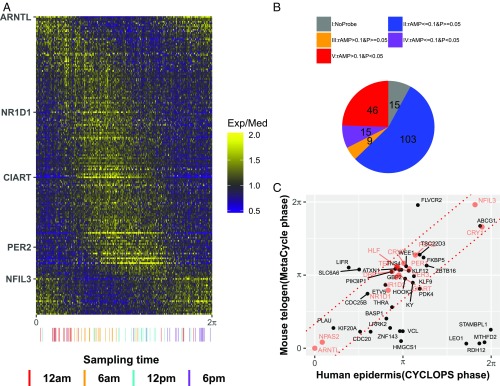
Fig. 3.
Population analysis of human epidermis data revealing conserved circadian transcriptional output. (A) Heatmap of median centered gene expression values for all human samples (n = 298) ordered by CYCLOPS phase and all significant circadian genes (n = 188). Clock genes that cover a range of circadian phases (e.g., ARNTL, NR1D1) are labeled on the y-axis. Samples with known sample acquisition times (n = 79) are shown along the x-axis. (B) All circadian genes in human epidermis. Groups are no mouse homolog (gray), low amplitude (rAMP ≤0.1) and not cycling (P ≥0.05) (blue), high amplitude (rAMP >0.1) and not cycling (orange), low amplitude and cycling (P <0.05) (purple), and high amplitude and cycling (red). (C) A comparison of high-amplitude cycling genes (n = 46) revealing strong phase conservation between mice and humans. Clock and clock-associated genes are shown in red. The predicted gene phases from CYCLOPS and MetaCycle were adjusted to ARNTL/Arntl phase (set to 0).