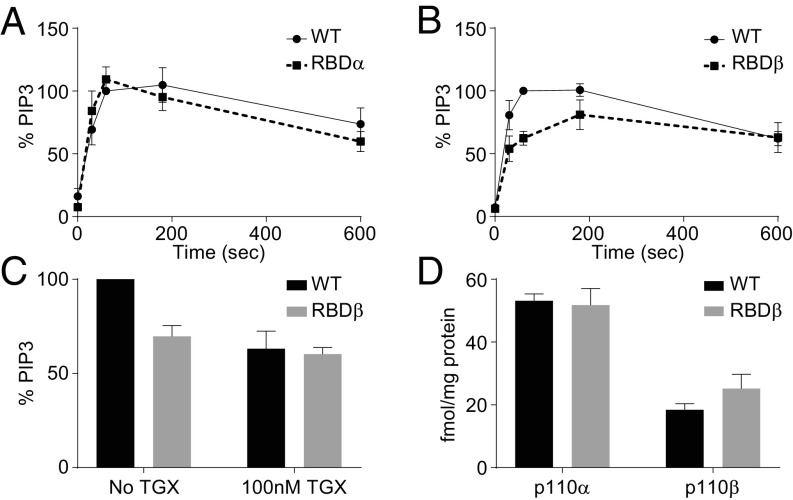
Fig. 8.
The RBD domain of p110β is necessary for PIP3 production but not for binding to activated PDGFRs. (A) PDGF-stimulated (PDGF-BB, 10 ng/mL, 1 min) PIP3 accumulation in MEFs expressing wild-type (WT) or Ras-insensitive p110α (RBDα). Results are expressed as percentage of the PIP3 levels in the respective, wild-type (WT) line. The data are means, ±SEM, from three experiments. (B) PIP3 accumulation in MEFs expressing WT or Rac/CDC42-insensitive p110β (RBDβ). Data are expressed as in A. The amount of PIP3 that accumulated in MEFs expressing RBDβ, but not RBDα, was significantly lower than their WT controls (P = 0.011, two-way ANOVA). (C) PIP3 levels in PDGF-stimulated (PDGF-BB, 10 ng/mL, 1 min) WT or RBDβ-MEFs in the presence, or absence, of TGX-221 (100 nM). Results are expressed as percentage of the PIP3 levels measured in the respective WT-MEFs in the absence of inhibitor. The data presented are means, ±SD, from three experiments, TGX-221 significantly inhibited the response in WT- (P = 0.03, ratio paired t test) and RBDβ-MEFs (P = 0.03, ratio paired t test). (D) The absolute amounts of p110α or p110β recruited to activated PDGFRs (PDGF-BB, 10 ng/mL for 1 min) in RBDβ- or WT-MEFs. The data are presented as means, ±SD, from three experiments, the amount of p110β associated with the activated PDGFRs was significantly lower than p110α (P = 0.00003 in WT and 0.0027 in RBDβ-MEFs; t test, two-sample equal variance, two-sided distribution); however, the association of neither p110α nor p110β was significantly changed in the context of the RBDβ mutant construct. The data underlying D is shown in SI Appendix, Fig. S15.