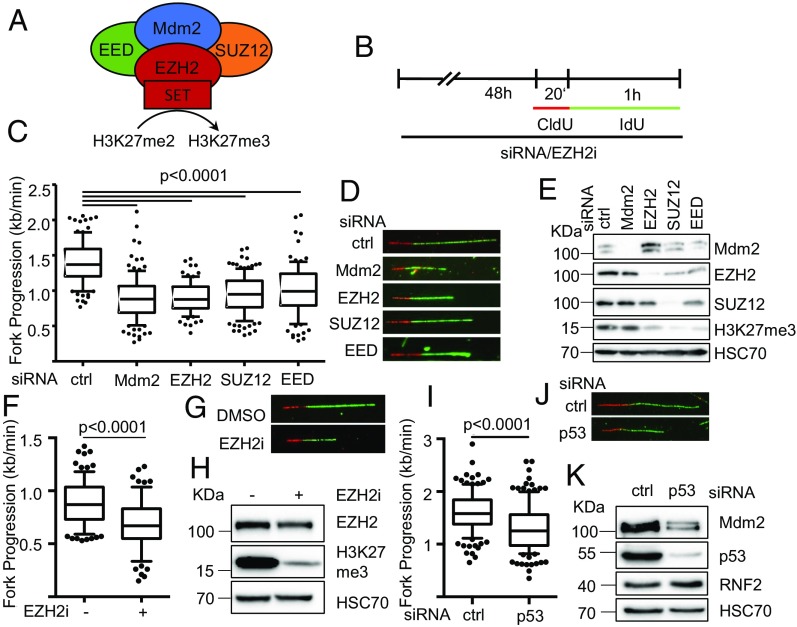
Fig. 1.
Compromised DNA replication fork progression upon depletion of Mdm2 or PRC2 members. (A) Schematic diagram of the main components of the PRC2 complex and its interaction partner Mdm2 that have been subjected to replication studies. siRNA transfections targeting Mdm2, EZH2, SUZ12, and EED as well as an inhibitor targeting the SET domain of EZH2 were used. (B) H1299 cells were transfected with siRNAs each against the targets Mdm2, EZH2, SUZ12, and EED or treated with the EZH2 inhibitor EPZ-6438 for 48 h and incubated with 5′-chloro-2′-deoxyuridine (25 μM CldU, 20 min) followed by 5′-iodo-2′-deoxyuridine (25 μM IdU, 60 min) as indicated. (C) Fork progression was determined from the track length of the second label (IdU; kilobases per minute) and displayed in a boxplot with 10–90 percentile whiskers. (D) Representative images of tracks of newly synthesized DNA were visualized by immunostaining of CldU (red) and IdU (green). (E) Cells were treated as in B and subjected to immunoblot analysis. Knockdowns were confirmed by staining for the target proteins as well as the target of active PRC2, i.e., trimethylated histone 3 (H3K27me3). (F) H1299 cells were treated with 5 μM of the EZH2 inhibitor EPZ-6438 for 48 h, followed by incubation with CldU and IdU as in B. Boxplot analysis of the fork progression during the time of IdU label with 10–90 percentile whiskers. (G) Representative labeled tracks were immunostained as described in D. (H) Cells were treated as in F and subjected to immunoblot analysis of H3K27me3 as a readout for inhibitor activity. SJSA cells were transfected with siRNA targeting p53 for 48 h followed by incubation with CldU (25 μM; 20 min) and IdU (25 μM; 60 min) for fiber analysis. (I) The fork rate of replication during the IdU label is displayed in boxplots with 10–90 percentile whiskers. (J) Representative images of CldU (red) and IdU (green) labeled tracks. (K) Immunoblot analysis of proteins after siRNA treatment described in I to confirm p53 depletion and protein levels of Mdm2 and RNF2 in response to p53 knockdown. In addition to the DNA fiber assays shown in this figure, replicates with the siRNAs used, alternative siRNAs, as well as in HCT116 p53−/− cells, are shown in SI Appendix, Fig. S1.