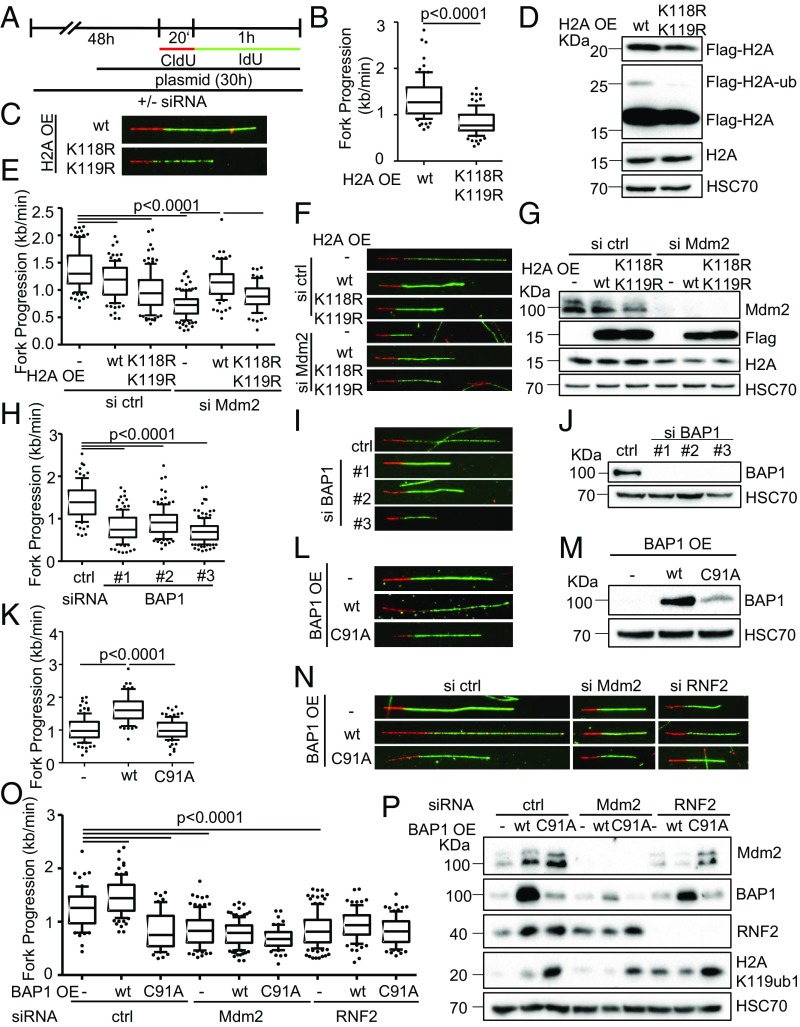
Fig. 5.
Dynamic changes in H2A ubiquitination are required for DNA replication. (A) H1299 cells were transfected with siRNA for a total of 48 h, and with plasmid DNA for 30 h before DNA fiber or immunoblot analysis. (B) Cells transfected with plasmids to express wild-type H2A (pcDNA3.1-Flag-H2A) or a mutant lacking the lysine residue that is ubiquitinated by Mdm2 and RNF2 (K119; pcDNA3.1-Flag-H2A-K118-119R) were assessed regarding their fork progression in the IdU label. (C) Representative images of fluorescently labeled tracks corresponding to experiment in B. (D) Immunoblot staining of cells treated as in B for Flag-tagged H2A expressed from plasmids. Note that the Upper band represents ubiquitinated H2A and is observed only with wild-type H2A. (E) Cells were transfected with siRNA targeting Mdm2 followed by ectopic expression of wild-type or mutant (K118-119R) H2A or an empty control plasmid. Fiber analysis was carried out 30 h post plasmid transfection using 50 μM CldU and IdU (20 and 60 min). (F) Representative labeled tracks of CldU (red) and IdU (green) of Mdm2-depleted cells with ectoptic H2A expression. (G) Immunoblot analysis to confirm Mdm2 depletion and the expression of Flag-tagged H2A plasmids. (H) H1299 cells depleted of BAP1 using three different siRNAs. Transfections were carried out for 48 h, followed by DNA fiber analysis to monitor replication fork progression. The track lengths of the IdU label are displayed in a boxplot. (I) Fluorescently labeled tracks of control and BAP1-siRNA transfected H1299 cells corresponding to H. (J) Immunoblot analysis of samples treated as in H to confirm depletion of BAP1 with siRNAs. (K) H1299 cells were transfected with plasmids expressing the deubiquitinating enzyme BAP1 and a catalytically inactive point mutant of BAP1 (C91A) for 30 h. Fiber analysis was carried out using 50 μM CldU and IdU for 20 and 60 min, respectively. (L) Fluorescent staining of CldU (red) and IdU (green)-labeled tracks corresponding to K. (M) Ectopic expression of BAP1 from plasmids was assessed by immunoblotting and staining with an antibody to BAP1 after 30 h of transfection. (N and O) Cells depleted of Mdm2 and RNF2 were transfected with plasmids encoding BAP1 as wild-type or a catalytically inactive mutant and subjected to fork progression analysis. Representative images of labeled tracks in N and fork progression rates in O. (P) Lysates of cells treated as in O were assessed for levels of Mdm2, RNF2, BAP1, as well as the substrate of the three enzymes, H2AK119ub1. Replicates to the fiber assays in this figure are shown in SI Appendix, Fig. S5.