Significance
The phenomenon of genetic balance is characterized by the finding that addition or subtraction of a single chromosome to the whole set is more detrimental than altering the dosage of the complete complement. The molecular basis of this principle has not been studied previously in a comprehensive manner. In this study, the set of all five trisomic chromosomes and a ploidy series of diploid, triploid, and tetraploid in Arabidopsis were examined for global gene expression modulations. The results indicate an impact of genomic stoichiometry on the landscape of gene expression, which has implications for how gene expression operates, the evolution of duplicate genes, and the underlying basis of quantitative traits.
Keywords: aneuploidy, polyploidy, trisomy, dosage compensation, gene balance hypothesis
Abstract
Changes in dosage of part of the genome (aneuploidy) have long been known to produce much more severe phenotypic consequences than changes in the number of whole genomes (ploidy). To examine the basis of these differences, global gene expression in mature leaf tissue for all five trisomies and in diploids, triploids, and tetraploids of Arabidopsis thaliana was studied. The trisomies displayed a greater spread of expression modulation than the ploidy series. In general, expression of genes on the varied chromosome ranged from compensation to dosage effect, whereas genes from the remainder of the genome ranged from no effect to reduced expression approaching the inverse level of chromosomal imbalance (2/3). Genome-wide DNA methylation was examined in each genotype and found to shift most prominently with trisomy 4 but otherwise exhibited little change, indicating that genetic imbalance is generally mechanistically unrelated to DNA methylation. Independent analysis of gene functional classes demonstrated that ribosomal, proteasomal, and gene body methylated genes were less modulated compared with all classes of genes, whereas transcription factors, signal transduction components, and organelle-targeted protein genes were more tightly inversely affected. Comparing transcription factors and their targets in the trisomies and in expression networks revealed considerable discordance, illustrating that altered regulatory stoichiometry is a major contributor to genetic imbalance. Reanalysis of published data on gene expression in disomic yeast and trisomic mouse cells detected similar stoichiometric effects across broad phylogenetic taxa, and indicated that these effects reflect normal gene regulatory processes.
The concept of genetic imbalance has been known for nearly a century, and originated from the finding that changing the dosage of individual chromosomes (aneuploidy) has a more detrimental effect on the phenotype than changing the dosage of the entire set of chromosomes (ploidy) (1–3). As molecular genetics developed, the simple assumption emerged that this phenotypic effect resulted from the varied genes showing a dosage effect. This is clearly the case for at least some genes; otherwise, there would be no impact. However, some evidence in maize and Drosophila indicated the presence of global genome-wide cascading modulations (4–8). By contrast, other studies have assumed that these disruptions of gene expression on numerically unaltered chromosomes in aneuploids are minimal (9, 10).
The fact that transcription factors and signal transduction components are typically dosage-sensitive (11–15), however, would suggest that their targets would be modulated regardless of the chromosomal location of the latter. Indeed, copy-number variants (CNVs) of transcription factors and signaling components often condition clinically recognized disease states in humans (16–18). Studies of experimentally produced chromosomal dosage changes can provide critical data that have implications for genetic control of gene expression and quantitative traits that are influenced by natural quantitative variation for regulatory components.
Furthermore, evolutionary genomics reveals a generalized pattern of selective gene retention after whole-genome duplication (WGD), with genes encoding members of macromolecular complexes, including transcription factors and signaling components, being maintained for longer periods of evolutionary time (19–26). Underrepresentation of duplications of the same classes of genes in populations as natural variation indicates a complementary pattern, illustrating that the genomic parameters of balance play out in selection in populations (17, 18, 22, 27). In other words, when genes involved in macromolecular complexes are out-of-register with their interactors, there are negative fitness consequences. This same evolutionary pattern has occurred in most taxa of eukaryotes, including yeast, protozoa, vertebrates, and especially the plant kingdom (28–30). While these evolutionary studies have been expanding in the past decade, there has been little information about how genomic balance affects gene expression. Arabidopsis is a good model for these types of studies because the evolutionary history of whole-genome duplications has been documented and a complete set of primary trisomies as well as a polyploid series are available for examination.
Results
Trisomies Show a Greater Spread of Modulation than Ploidy.
An RNA-sequencing (RNA-seq) study of mature leaf tissue was performed using each of the five trisomies of Arabidopsis thaliana, ecotype Columbia, with comparison with the normal diploid. In concert, a ploidy series of diploid, triploid, and tetraploid was investigated. One approach for examining the expression trends was to produce ratio distributions of experimental and control read counts. This type of analysis provides a topology of narrow-range modulations of mRNA expression that comparisons of individual genes between genotypes do not, because all genes are used to detect trends, including multiple groupings. Normalization was performed using an External RNA Controls Consortium (ERCC) spike-in to total RNA followed by polyA RNA isolation rather than to endogenous gene expression to detect any transacting effects of aneuploidy on global gene expression. Exogenous spike-in to total RNA provides a robust procedure for analysis of global modulations of gene expression (31). For this type of analysis, read counts were averaged across biological replicates. Ratios between the compared genotypes were generated for every expressed gene and then plotted as a ratio distribution. No relative change in expression is designated by the ratio 1.00. Statistical determinations of deviation of the distributions from normal (SI Appendix, Table S1) or to compare two distributions for differences (SI Appendix, Table S2) used Kolmogorov–Smirnov (K-S) tests of significance. Bartlett’s test was performed to compare variances across groups (SI Appendix, Table S3). This approach was further cross-validated by determining significant fold change of individual gene ratios that differ from no change (1.00) in t tests (see Materials and Methods for details) and plotting those values in scatter plots. Further validation came from selected sampling of gene expression differences by quantitative PCR using an external spike-in. Together, this approach provides a highly robust method involving multiple cross-validation to determine both cis and trans effects of aneuploidy and ploidy on gene expression.
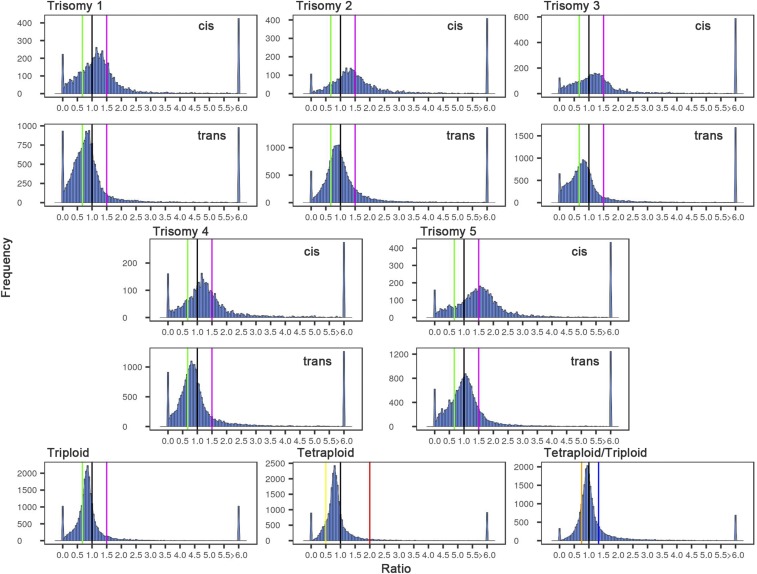
The ratio distributions of the primary trisomies show a spread across a landscape of effects with each being distinct, whereas the ploidy comparisons show tighter distributions (Fig. 1). The patterns for the trisomies are broken into those genes that are present on the varied chromosome (cis) versus those that are in the remainder of the genome (trans). For each trisomy, the distributions comparing cis and trans are clearly different but there is a range in the effects observed. The cis effects show peak groupings between that typical of a dosage effect (∼1.50-fold) and no difference (dosage compensation) (1.00) but also a spread that extends above and below these levels. The greatest peak in cis for trisomies 1 and 4 are closer to dosage compensation than dosage effect. For trisomies 2 and 3, the major cis peak is intermediate. Trisomy 5 shows a close approximation to a generalized dosage effect in cis.
Fig. 1.
Ratio distributions of gene expression in each trisomy and ploidy compared with diploids. The sequencing reads from RNA-seq, analyzed as described in Materials and Methods, were averaged for the biological replicates. For each expressed gene, a ratio of the averaged read counts in the respective experimental (trisomy or ploidy) genotype was made over the read counts in the diploid control. These ratios were plotted in bins of 0.05. The x axis notes the value for each bin, and the y axis notes the number of genes per bin (frequency). For the five trisomies, genes were partitioned into those encoded on the varied chromosome (cis) versus those encoded on the remainder of the genome that were not varied in dosage (trans). A ratio of 1.00 represents no change in the experimental genotype versus the diploid. A ratio of 1.50 represents a gene-dosage effect in cis, whereas 1.00 represents dosage compensation. A ratio of 0.67 represents the inverse ratio of gene expression in trans. These ratio values are demarcated with labeled vertical lines in magenta (1.50) and green (0.67). The triploid and tetraploid ploidy series were analyzed in the same manner for all expressed genes. The vertical demarcations in this case correspond to the respective direct or inverse relationship of the ploidy comparison in magenta and green and red (2.00) and yellow (0.50), respectively. The tetraploid/triploid comparison was generated by producing the respective ratios and plotting the distribution with the direct and inverse relationship depicted with vertical lines in blue (1.33) and orange (0.75). Each comparison is labeled in the respective panel.
The trans effects in the trisomies also exhibit a wide spread, with the most common trend being a reduced expression in the trisomy compared with the normal diploid, with a ratio of 1.00 being defined as no modulation. The fact that the cis and trans effects are modulated coordinately to some extent (depending on gene functional groups; see below) indicates a common influence of the aneuploid state, in general, on gene expression on both the varied and unvaried chromosomes. Nevertheless, regardless of the global modulations, the cis and trans distributions remain distinct (SI Appendix, Fig. S1; t test of means, P < 0.0002; Wilcoxon test, P = 0.008). This generalized observation indicates a minimal level of feedback or buffering effects as the basis of compensated genes, but rather the lack of a significant dosage effect is usually the result of the combination of an increase in gene dosage modulated by a down-regulation (32). Scatter plots testing the significance of the cis and trans ratios complement the distributions (SI Appendix, Fig. S2). Quantitative PCR of specific genes for relative expression levels in selected trisomies and ploidy comparisons was conducted and the results are in agreement with the individual gene placements in the distributions (SI Appendix, Fig. S3).
The ratio distributions comparing ploidies of triploid with diploid and tetraploid with diploid show much less spread, and in each case the sharp peak is near 1.00 but in each case slightly below (Fig. 1). Cell size in a ploidy series of wild-type Arabidopsis is correlated with the number of copies of the genome (33, 34). The similarity of the shape of the distributions and its center not far removed from 1.00 suggests that the cells increase in size from diploid to triploid to tetraploid, with a correlated change in overall gene expression per cell. The fact that the ploidy distributions are near-normal but the peaks are slightly less than 1.00 might indicate that the assayed genes are diluted in the transcriptome, potentially by slightly nonlinear expression compared with ribosomal RNA (the major component of total RNA to which the exogenous ERCC spike-in was added). If the two higher ploidies were identical to each other but with both being different from the diploid (if the latter is an outlier in the ploidy series), the tetraploid/triploid ratio distribution would approach a normal distribution surrounding 1.00. However, the magnitude of the reduction scales with ploidy, because the tetraploid/triploid ratio distribution has a peak slightly below 1.00 but less so than in the tetraploid/diploid distribution (Fig. 1).
Nevertheless, any relative differences for expression of protein-encoding genes between ploidies would be reflected in the ratio distributions, and thus there appears to be much fewer such differences across the ploidy series than in the various trisomies. The SDs of the means of the ploidy distributions are uniformly of lesser magnitude than those of the trisomies (SI Appendix, Table S1). Also, the trans distributions for each of the five trisomies were compared with the triploid and tetraploid distributions in K-S tests. These comparisons showed that the probability that they are the same is essentially zero (SI Appendix, Table S2). Statistical comparisons of the variances of the trans distributions of the five trisomies with ploidy distributions in Bartlett’s tests showed highly significant differences (SI Appendix, Table S3), substantiating a greater disruption of gene expression in aneuploidy than in polyploidy. These molecular findings parallel the phenotypic relationships long known in such comparisons (35).
Trisomies and the ploidy series have some phenotypic and growth differences compared with diploid controls (36). A further advantage of the ratio-distribution analysis is that it has the power to detect whether there are differences in cell type and developmental gene expression in the experimental genotypes relative to the diploid. If these differences are substantial, the outliers in the distributions (ratios >6.00 and near 0.00) would be heavily populated by cell- and developmentally specific genes. However, examination of the distributions indicates that these outliers are minor contributors to the total, suggesting that this concern is unwarranted for sampling of the leaf tissue. To the extent that such effects do occur, the ratio-distribution approach separates any such effects from the others occurring in the expression landscape. Indeed, we did not observe overrepresentation of cell- and developmental related categories in a Gene Ontology enrichment analysis for biological processes of the outliers (SI Appendix, Table S4). Partitioning the genes into arbitrary low, medium, and high expression followed by generating ratio distributions shows that the outliers are predominantly in the low-expression comparisons. This analysis suggests that small increases or decreases of ratios due to low read counts in the numerator or denominator can shift the ratio out of the central distribution (SI Appendix, Fig. S4).
Genes in Different Functional Groups Show Diverse Responses to Aneuploidy.
Genes were partitioned into functional groups to determine whether there were any specific responses to aneuploidy and ploidy. As noted above, evidence from evolutionary genomics indicates a preferential retention following WGD in Arabidopsis of many genes encoding transcription factors, signal transduction components, ribosomal proteins, and the subunits of the proteasome (21, 23, 28)—all participating in macromolecular complexes. Transcription factors (Fig. 2 and SI Appendix, Fig. S5) and signaling components (SI Appendix, Fig. S6) have trans distributions with an affected peak in each case near the inverse level (0.67) or below. Signal transduction genes have similar distributions compared with all genes (SI Appendix, Table S2). The distributions for transcription factors for each trisomy are highly significantly different from the distributions for all genes (SI Appendix, Table S2).
Fig. 2.
Ratio distributions of expression of transcription factors in the noted comparison. Analysis was conducted as described in Fig. 1 using only annotated transcription factors as described in Materials and Methods.
Genes encoding ribosomal structural proteins (Fig. 3 and SI Appendix, Fig. S7) and the proteasomal proteins (SI Appendix, Fig. S8) are much less modulated, indeed much less so than the trend for all genes for all five trisomies, as statistically verified by K-S and Bartlett’s tests (SI Appendix, Tables S2 and S3). The mRNAs for the structural components of the ribosome constitute ∼3 to 4% of the total pool of mRNAs in this study but are distinct in their reaction to trisomy. In general, they are less inversely affected than all genes, although a small fraction is negatively modulated. Trisomy 2 has a strong positive effect in trans on some ribosomal structural genes, but all five trisomies show a bipartite effect in trans (Fig. 3). Such imbalance of ribosomal proteins potentially contributes to phenotypic effects (37). However, because trisomy 2 does not have an unusual ratio distribution for other classes of genes, or for all genes, this response of the mRNAs for ribosomal proteins does not appear to affect the overall analyses. The ribosome and proteasome groupings also show less spread in the distribution in the ploidy comparisons (Fig. 3 and SI Appendix, Figs. S7 and S8; p ∼ 0, SI Appendix, Table S3). Lastly, an analysis of stress-associated genes was performed to test the proposition that aneuploidy might induce genes in that category (38) (SI Appendix, Fig. S9). Examination of each trisomy and across all trisomies showed a similar pattern of narrow-range modulations of the stress-associated genes to the total gene distributions. Importantly, however, the stress genes do not populate the outlier peaks to any great extent, indicating that there is no generalized induction of their expression to higher or lower extremes in these types of aneuploids (SI Appendix, Table S4).
Fig. 3.
Ratio distributions of expression of genes encoding the structural components of the ribosome. Analysis was conducted as described in Fig. 1 using only annotations for ribosomal structural proteins as described in Materials and Methods.
For nuclear genes whose products are targeted to the organelles, which must coordinate their own expression with that of the nucleus (39), the chloroplast (Fig. 4 and SI Appendix, Fig. S10), and the mitochondrion (SI Appendix, Fig. S11), there are stronger reductions in each trisomy than for all genes (SI Appendix, Table S2). The major peak for the genes encoding plastid-targeted proteins in particular closely tracks the inverse level in trisomies 2 to 4, and in 1 and 5 spans the range between the inverse level and no change for the trans expression. The distributions for cis genes with encoded proteins targeted to the chloroplast correspondingly exhibit peaks at dosage compensation for trisomies 2 to 4. The tissue studied here is mature leaves that are actively photosynthesizing and, indeed, this functional class comprises 43% of the diploid mRNA transcriptome. This strongly expressed group of genes is primarily inversely affected. For the peroxisome (SI Appendix, Fig. S11), which does not contain DNA and hence no organelle-synthesized components, a similar modulation to that found with other organelles occurred.
Fig. 4.
Ratio distributions of expression of genes encoding proteins targeted to the chloroplast. Analysis was conducted as described in Fig. 1 using only annotations for chloroplast-targeted proteins as described in Materials and Methods.
DNA Methylation Status in Trisomies.
DNA methylation has been reported to be increased across the genome in human trisomic individuals (40–48). Methylation states in protein-coding sequences were determined for each Arabidopsis trisomy and compared with the diploid (49) (Materials and Methods and SI Appendix, Table S5). In each trisomy, four states of methylation were assessed: unmethylated, mCG methylation in gene bodies, and methylation in mCHG and mCHH nucleotide groups. The largest effect was found for trisomy 4, in which a notable shift between unmethylated and CG gene body methylation to CHH methylation was observed (SI Appendix, Table S5). To test whether this change in DNA methylation is related to modulations of gene expression, the genes with an acquired CHH methylation state in trisomy 4 were compared with those with no change, but there was not a dramatic shift in expression to the outlier peaks of the ratio distributions (SI Appendix, Fig. S12). Although CHH methylation has been associated with transcriptional repression, especially for transposable elements (50), the unusual association with gene bodies in trisomy 4 observed here does not have an obvious effect on gene expression.
We also determined the expression ratio distributions of unmethylated genes and CG gene body methylated genes for all trisomies. Those with gene body methylation, which as a group generally represent housekeeping genes (49), show distributions with less effect compared with those of unmethylated genes (SI Appendix, Fig. S13), which are highly significantly different for all trisomies (SI Appendix, Table S2) and have significantly different variances of the distributions compared with unmethylated genes (SI Appendix, Table S3). Trisomy distributions of CHH methylated genes were scattered without discernible trends, but this might be due to the low number of genes analyzed (SI Appendix, Fig. S13).
Polycomb Complex-Associated Genes.
The Polycomb repressive complex has a regulatory role that reduces the expression of associated genes with a genetic behavior that is typically dosage-sensitive (51). Thus, it is of interest to determine whether they are capable of being modulated by trisomy, given that the plurality of effect of the latter is also a reduced expression. There is a spread of modulations but the trend is toward less expression in the aneuploids (SI Appendix, Fig. S14), despite the fact that Polycomb-associated genes even in the diploid are in a state of lowered function (51). Therefore, Polycomb repression does not eliminate the effect of genomic imbalance. The results suggest that the impact of trisomy is independent of the Polycomb complex and is additive.
Genetic Imbalance Causes Discordance Between Transcription Factors and Their Targets.
Selected transcription factors (TFs) and their predicted targets have been defined in expression networks (52). The Arabidopsis thaliana Regulatory Network (AtRegNet) describes the relationship of TFs and their direct target genes as to whether the targets are induced or repressed by a given TF (52). We compared the expression of TFs with that of their targets for each trisomy and ploidy change. TFs and their targets that are known were examined via ratio distributions in the trisomy data when the TF-encoding genes were on the varied chromosome (SI Appendix, Fig. S15). We also analyzed the distributions of TF expression and their targets that are not on the varied chromosome but that are modulated in expression in trans (SI Appendix, Fig. S15). These cases would represent those in which the trisomy changes the expression of an unlinked transcription factor, which in turn modulates its targets in a cascade of effects. From these distributions, we used only genes showing a significant differential expression generated by empirical analysis of digital gene expression data in R (edgeR) (53, 54) versus the diploid control (Materials and Methods) to compare with previously published interactions (SI Appendix, Table S6). Some trisomies show a considerable discordance between the positive or negative effects predicted from the network data and the aneuploidy response when identifying the expression of TFs in cis and their predicted targets. Across all five trisomies there is a range of discordance from 42 to 62% between the TF–target network relationship and the response in trisomy. Of course, the identified targets of transcription factors could be affected by other factors modulated by trisomy from the specific transcription factor examined, and the specified networks are likely to be variable under different conditions. In the whole-genome ploidy series there are few significant effects, because there is a lack of relative change in gene expression, as described above. The comparison of aneuploidy and ploidy illustrates the disruptive “dominant-negative” impact of altered genomic stoichiometry on gene expression when transcription factors or other regulatory molecules are varied in isolation from their interactors.
Impact of Genomic Imbalance in Other Phyla.
There have been suggestions in the literature that the modulations across the genome in aneuploids of plants and Drosophila were specific to these taxa (9, 10). However, it seems likely that the kinetics of cellular reactions would be universal. Studies in yeast disomics (55) and selected mouse trisomic cell lines (56) have previously examined gene expression on the genomic level in microarray experiments. The data were analyzed previously by normalizing the expression of genes from the varied chromosome by the unvaried portion of the genome under the assumption that there are no general modulations across the genome. Based on the finding here that the most prominent effect in hyperploidy in trans is a down-modulation, the previous approach would inflate the apparent expression of the varied chromosome in the treated data and obscure to some degree the effects in trans.
We therefore reexamined those data producing ratio distributions of cis and trans genes independently. These data have no exogenous normalization control, so modulations in the same direction from no change will be muted in relation to the strength of the effect and thus stronger biologically than the ratio distributions might indicate (57). Nevertheless, modulations were detected, with each hyperploid being distinctive (SI Appendix, Figs. S16 and S17). Some aneuploids show little modulation across the genome while others exhibit varying degrees of down-regulation, as expected from different genic content per chromosome and as also found in the present study of Arabidopsis trisomies. Ratio distributions of technical replicates in the yeast study showed little spread from 1.00 (SI Appendix, Fig. S16), illustrating the high quality of the data acquisition. This comparison illustrates that the deviations from normal that occur in the aneuploid/euploid comparisons reflect biological reality, which is supported by high significance in Bartlett’s test comparing disomic distributions with those of technical replicates of haploids (SI Appendix, Table S3). When modulations in trans are evident, it should be noted that the spread from no change (1.00) is greater for the yeast disomies than for the mouse trisomic cells and for Arabidopsis (this study) and Drosophila trisomies (7, 8), consistent with the degree of genomic imbalance impacting the magnitude of modulation. The analyses here indicate the commonality of aneuploidy effects on gene expression and their major trends across phyla, and demonstrate the need to reevaluate conclusions from yeast and mammals that rely on the assumption of little to no effect across the genome.
Discussion
The phenomenon of genetic balance has been studied for nearly a century (1, 3), but the underlying basis remains to be defined. In this study, gene expression on the RNA level was examined in all five trisomies of Arabidopsis, which cover the entire genome, and in a polyploid series of diploid, triploid, and tetraploid using mature leaf tissue. There is a generalized greater spread of modulations in aneuploids than in the ploidy series. The plurality effect in trisomy is a reduced expression, but there is clearly a complicated topology of both increases and decreases compared with the normal diploid control. There are differences in response to aneuploidy depending on gene function, with ribosomal and proteasomal components showing distinct effects compared with transcription factors, signaling components, and organelle-targeted genes, as examples. The functional group with the highest expression in the experimental material, genes encoding plastid-targeted proteins involved in photosynthesis, was primarily inversely affected in the trisomic genotypes.
Throughout eukaryotes there is a generalized correlation between ploidy level and cell size (58–60). Because our normalization involved an exogenous spike-in to total RNA, which correlates with cell size in Arabidopsis (60), the ratio distributions for the ploidy series should be interpreted with the view that there is a greater amount of total RNA per cell with increasing ploidy. Thus, the ploidy series ratio distributions do not reflect the increasing overall transcriptome but instead reveal the relative changes that occur, which were few.
In contrast, extensive anatomical studies of cell size in Datura trisomies did not identify any general trend but some instances of specific cell types that were decreased or increased in size or number (58, 61). As noted above, the ratio-distribution analysis can detect major differences in cell type-specific expression between trisomies and normal, but little evidence for such was found. In various studies, cellular observations of Arabidopsis diploids, tetraploids, hexaploids, and octoploids revealed the canonical increase in size for ploidy (59, 60) but no perceptible cell size differences for any of the five trisomies has been documented compared with the normal diploid (59, 62), providing no evidence for a generalized increase or decrease in total RNA per cell in trisomies. In maize, total ribosomal RNA (the major constituent of total RNA) per DNA was measured in a large number of trisomies and other aneuploidies with no case of altered relative amounts being found (6).
If we consider a scenario in which the total RNA transcriptome is modulated independently of the total mRNA transcriptome size, which in a ploidy series of Arabidopsis does not occur but instead the two correlate more closely with each other than with the genome copy number (60), an increase or decrease in total RNA would produce ratio distributions with uniform deviations from the diploid. However, the ratio-distribution analysis of the trisomies detects significantly different ratios that are split in their response either positively, negatively, or with no change (compare functional groups as gene body methylated genes versus all genes or the split of significant positive and negative ratios within a single functional group, such as the genes encoding ribosomal proteins, among others) that cannot result from a significant disconnect between the total mRNA transcriptome and the total RNA transcriptome having an effect on the analysis. Whereas the plurality peaks in cis and trans track to some degree in all trisomies (SI Appendix, Fig. S1), there is a spread in all distributions that indicates this is not due to a discordance between total mRNA and total transcriptome sizes but rather to coordinate modulation of subsets of genes. Moreover, the considerable dissimilarities between aneuploid expression and regulatory networks in trisomies, but not in the ploidy series, further support this conclusion. Given these observations and the lack of evidence for any consistent cell-size change, we believe the analysis is an accurate reflection of the types of modulations occurring in the Arabidopsis trisomies.
Studies of gene expression in aneuploids have come to different conclusions about the prevalence of dosage effects or compensation of the varied genes (57). These determinations are complicated by the narrow range of expression difference, normalization procedures, the criteria of classification for dosage effect or compensation, and the tendency to view compensation necessarily as a chromosome-wide effect. Our data show evidence for a range of effects that in some cases exceeds the limits of dosage effect or compensation. Critically, there are widespread changes of gene expression across the whole genome, with decreased expression being the most prominent modulation caused by hyperploidy.
Several previous studies found increased DNA methylation in human trisomy 21 individuals as well as for trisomies 13 and 18 (40–48), and methylation might play a role in genome expression dominance in allopolyploids (63). In this Arabidopsis study, a few genes shift methylation status between genotypes, particularly for trisomy 4, indicating that trisomy can affect the methylation status in the genome. However, there is not a generalized effect of aneuploidy on gene body methylation. With regard to gene expression, the trisomy response of genes with gene body methylation is less than genes that are unmethylated.
Although presaged by phenotypic data showing a predominance of reduced expression in segmental trisomies in the classical genetics literature (64), the plurality finding that hyperploidy reduces gene expression is counterintuitive to the generally accepted view that gene expression in eukaryotes operates predominantly positively, which is supported by transcription factor/target gene network data. However, the results obtained here follow the predictions of kinetic considerations of how macromolecular complexes, typically involved in gene expression, assemble (65–69). When changing the stoichiometry of different types of bridge subunits relative to others, the assembly of the whole is compromised. Of particular interest for the present study would be increased relative bridge subunit concentration, which might occur in trisomies if the bridge molecule is increased in expression relative to others in the complex. The altered stoichiometry could arise from a gene-dosage effect of the varied chromosome or from trans modulation, depending on the specific circumstances. However, regardless of the modulation in the cell, the cis and trans expression shows an altered stoichiometry (SI Appendix, Fig. S1). In essence, the system operates as a multilocus dominant negative on gene expression. There are many nonlinear steps between varying the dosage of a gene and the steady-state level of its product. However, those cases that approximate such a relationship would be reflected in the observed results comparing aneuploidy and polyploidy as described here.
The data are consistent with the concept that gene regulatory processes follow the same principles of genetic balance that have been known on the phenotypic level for most eukaryotic organisms and thus are the ultimate underlying basis of these organismal effects (5). When dosage-sensitive transcription factors are varied, they modulate their targets. Of course, the effects will be modified by differential rates of synthesis and degradation of RNA and their encoded proteins, among other considerations of cellular metabolism and homeostasis (69, 70). Dosage sensitivity has been correlated with intrinsically disordered domains in proteins that facilitate multiple interactions, including for transcriptional and signaling proteins (71, 72). The mRNA and protein levels of these dosage-sensitive genes in general have tighter relationships than other functional classes (71, 72). Indeed, the comparison of the hyperploids with the whole-genome ploidy series, where there is less modulation, suggests that the change in quantity of some transcription factors and signaling components in isolation of the remainder of the genome can impact target gene expression. A transcription factor can be modulated in trans and in turn can modulate its target genes in trans, forming a cascade of effects across the genome.
Using databases that have connected transcription factors with their targets and comparing those patterns with the response in trisomy demonstrate a large portion with discordance, although these determinations are complicated by the fact that multiple regulatory factors could be modulated by the whole-chromosome trisomy. This is illustrated by comparing the expression distributions of transcription factors and their targets, which show considerable departure from those expected from network data, although the trisomic effects include many such pairings that are, in fact, concordant. Indeed, a mixed spectrum would be expected if the interaction context of any particular quantitative change of a TF is disruptive or not. As noted above, TFs are also regularly modulated in trans by trisomy and in turn affect their target genes, illustrating the complicated, connected effects that occur in aneuploidy.
The stoichiometric impact of transcription factors as a reflection of the kinetics of gene expression suggests that the responses seen in aneuploidy should be generally conserved among various organisms. Previous results in Drosophila trisomies (7, 8, 73–75) indicate global modulations across the genome with a spread of effects but with a predominant reduced expression for many genes. The effects could potentially be more pronounced in Drosophila because there is no readily discernible WGD in its evolutionary history (76), whereas the other taxa examined, including yeast and vertebrates, all have them in their ancestry (19, 22, 25, 29). Residual regulatory gene duplication might ameliorate the effective imbalance in those cases, as is known phenotypically for aneuploids in higher ploidy species (77). Reexamination of data from yeast (55) and mouse cell (56) hyperploids indicates a similar response to that observed in Arabidopsis, illustrating the commonality of this effect across taxa, which is also found in the literature for humans (78–80).
The generalized effect across phyla has implications for sex-chromosome evolution, which often results in a karyotype that on the surface resembles aneuploidy. The newly evolved sex chromosomes must accommodate and counteract the trans effects of regulatory mechanisms or maintain the dosage-sensitive regulators on the diverging homologs (81, 82). In mammals and birds, dosage-sensitive regulatory genes have been maintained on the homologs destined to become sex chromosomes (81, 82) as one means to prevent the global types of disruptions of expression noted in the aneuploids in this study. This conservation based on gene function parallels the retention of similar classes of genes following WGD across many taxa (28, 29) and illustrates the selective pressure to maintain the stoichiometry of regulatory mechanisms. Furthermore, X-linked dosage-sensitive genes involved in large protein complexes in mammals often escape X inactivation as a means of coordinating their levels with autosomal components (83). Nevertheless, imbalanced sex-chromosome genotypes in humans exhibit expression modulations across the genome (80).
Cancer cells are typically highly aneuploid. The stoichiometry of the gene regulatory machinery likely leads to complex and extensive modulations of gene expression that can affect the viability and growth characteristics of the cells. A study of karyotypes of a large number of cancer cell lines shows that the X chromosome and the larger autosomes regularly evolve to a particular genomic balance (84). The application of stoichiometric considerations to an understanding of cancer cells is suggested by our study.
The effects of these experimental changes in gene dosage indicate that natural variation in the quantity of individual transcription factors and signaling components would contribute to an intricate web of stoichiometric determinants of multigenic quantitative traits (6, 11, 37, 68). Copy-number variation on the individual regulatory gene level is suggested by our study to influence gene expression across the genome and contribute to quantitative trait variation. Just as multiple trisomies can influence overlapping sets of target genes, CNVs are suggested to do so as well and contribute to a complex, interacting, and nonlinear control of quantitative characters in eukaryotes. Our results not only illustrate the complicated spectrum of gene expression changes in aneuploidy but also have implications for how the regulatory stoichiometry influences genetic traits in general.
Materials and Methods
Production of Primary Trisomies.
To screen for primary trisomies of all five chromosomes of A. thaliana (2n = 10), we used a set of related transgenic lines (derived from the A.J.M.M. and M.M. laboratory strain of A. thaliana ecotype Col-0) in which each respective chromosome is marked with a fluorescent tag (85, 86). Diploid and tetraploid plants (produced by colchicine treatment) from each tagged line were crossed to generate triploid progeny. Seeds of the diploid and tetraploid fluorescence-tagged lines are deposited in the Arabidopsis Biological Resource Center, Ohio State University under ABRC nos. CS71713 to CS71722 and are available also upon request from the A.J.M.M. and M.M. laboratory. The triploids were allowed to self-fertilize, resulting in a swarm of aneuploids in the progeny population (87). Primary trisomies were identified by screening root nuclei of seedlings for three fluorescent dots (63× objective on a Zeiss Axioplan or Leica TCS LSI-III microscope), indicating the presence of three copies of a specific tagged chromosome. These plants, which could be either trisomic for the fluorescence-tagged chromosome, triploids, or more complex aneuploids (87), were saved and grown to maturity in soil. Three rosette-leaf samples were collected, frozen in liquid nitrogen, and stored at −80 °C. Upon flowering, immature pistils were harvested and used to prepare and count metaphase chromosomes as described previously (87). Only plants containing 11 chromosomes, indicating a trisomic state, were retained for further analysis. One leaf sample was used to prepare DNA for comparative genome hybridization (87) to confirm a specific trisomic condition and overall genome integrity. The other two leaf samples were used to prepare RNA and DNA for transcriptome and DNA methylome analysis, respectively. The chromosome constitutions of diploid, triploid, and tetraploid plants used in the transcriptome and methylome analysis were confirmed in the same manner.
Nucleic Acid Isolation.
Total RNA was isolated from rosette leaves using a Plant Total RNA Miniprep Purification Kit and RNA lysis solution B (GeneMark). Total DNA was isolated from rosette leaves using a Wizard Genomic DNA Purification Kit (Promega).
qRT-PCR.
Total RNAs were treated with RQ1 DNase (Promega) according to the manufacturer’s instructions. cDNA was synthesized using the Transcriptor First Strand cDNA Synthesis Kit (Roche) following the manufacturer’s protocol using an oligonucleotide d(T) primer and 600 ng of total RNA containing 2 µL 100× diluted ExFold ERCC controls (Life Technologies) as an internal control. Quantitative RT-PCR was performed using a 7500 Real-Time PCR System (Applied Biosystems) with the program recommended by the manufacturer using 1 µL cDNA as a PCR template and SYBR Green PCR Master Mix (Applied Biosystems). At least three technical replicates were carried out for each sample. Error bars indicate SE. Primer sets are shown in SI Appendix, Table S7.
Whole-Genome Bisulfite Sequencing.
Genomic DNA (100 ng) was sheared into fragments of 100 to 300 bp with a Bioruptor (Diagenode). Fragmented DNA was end-repaired, A-tailed, and ligated with premethylated adaptors. After size selection, adaptor-ligated DNA was treated with sodium bisulfite using a MethylCode Bisulfite Conversion Kit (Invitrogen) according to the manufacturer’s protocol. Bisulfite-converted DNA was amplified by PCR and sequenced with Illumina NextSeq 500 sequencers using the paired-end 150-cycle protocol.
RNA-Seq Library Construction.
Total RNA was quantified using the Qubit RNA BR Assay Kit (Invitrogen). Four micrograms of total RNA was spiked with ERCC ExFold RNA Spike-In Mixes (Thermo Fisher Scientific) according to the manufacturer’s protocol. RNA from diploid samples was spiked with Mix1 and others were spiked with Mix2. Spiked total RNA was used for library construction with a TruSeq Stranded RNA-Seq Library Prep Kit (Illumina). Briefly, polyA RNA was selected with oligo-dT beads. The first-strand cDNA was synthesized with reverse transcriptase, dNTP mix, and random primers. The second-strand cDNA was synthesized with dUTP mix. After end repair and A tailing, the DNA fragments were ligated with adaptors. After PCR amplification, the libraries were sequenced with Illumina HiSeq 2000 sequencers using the paired-end 100-cycle protocol. There were three biological replicates for the diploid and tetraploid and two biological replicates for the triploid and each of the five trisomic samples.
The traditional procedure to measure gene expression is calculating RPKMs (reads per kilobase per million mapped reads) for single-end RNA-seq or FPKMs (fragments per kilobase per million mapped reads) for paired-end RNA-seq. Because the DNA copy number is different in each aneuploid sample, RPKMs or FPKMs might be biased. To create a standard baseline measurement of gene expression, we added ERCC ExFold RNA Spike-In Mixes (Thermo Fisher Scientific), which are preformulated sets of 92 polyadenylated transcripts, to total RNA before polyA selection. Therefore, the exogenous RNA standards undergo all library construction steps, and reflect the performance of endogenous transcripts more closely.
After sequencing, mRNA reads were aligned to both the Arabidopsis reference genome TAIR10 (The Arabidopsis Information Resource no. 10) and ERCC sequences using TopHat2 (88), and the raw read counts (the number of reads mapped to the reference genome) were generated using Cuffdiff, which estimates the number of fragments that originated from each transcript (89). Meanwhile, the read counts mapped to ERCC sequences for all samples were generated. Then, the read counts of genes were normalized to generate a normalized count per million table using ERCC counts as a size factor via edgeR (53).
Statistical Analysis.
P values of normality check were generated by the Anderson–Darling test, Cramer–von Mises test, or Lilliefors test (SI Appendix, Table S1). P values used for similarity determinations between two ratio distributions were performed by the Kolmogorov–Smirnov test (SI Appendix, Table S2). χ2 goodness-of-fit tests were performed to examine if the number of genes with one category of methylation (e.g., gbM) in the diploid is significantly different from that in tetraploid, triploid, or trisomies (SI Appendix, Table S8). Bartlett’s test was performed to test if variances of distributions (with outliers—ratio >6 or <1/6—excluded) are equal across different groups (SI Appendix, Table S3).
Methylation Analysis.
For the methylation analysis, the methods described (49) were followed. In brief, the bisulfite sequencing data were filtered by requiring at least three reads of coverage at each cytosine. Filtered sites were then mapped against the Arabidopsis reference genome TAIR10 (https://www.arabidopsis.org/download/, October 26, 2017). Only reads mapped to the coding sequences of the primary transcript were retained (SI Appendix, Table S9). Methylation rates of mCG(gbM), mCHG, or mCHH for each gene were computed. Genes with no reads after filtering were classified as unknown. Afterward, a binomial test was applied to each gene for each sequence context, and q values were calculated by adjusting P values by the Benjamini–Hochberg false discovery rate (FDR). We classified genes into mCG(gbM), mCHG, or mCHH according to the criteria as described (49).
Ratio Distribution and Scatter Plots.
For generating ratio distributions, we filtered out genes with 0 read counts when combining experimental and control. Means of normalized counts of biological replicates of each treatment group (trisomy 1 to 5, triploid, or tetraploid, respectively) and the control group (diploid) were computed. The ratio was generated by dividing the mean of treatment counts by the mean of control counts. Then, ratio-distribution plots were generated with the ggplot2 package in the R program (90). The x axis of the plot demonstrates the ratio of treatment to control, whereas the y axis shows the number of genes per bin.
Student’s t test was performed to test for the significance of whether the mean log2 ratio of each treatment group (trisomy 1 to 5, triploid, or tetraploid, respectively) to the mean log2 control group (diploid) differed from 1.00. Genes with less than or equal to five counts when combining experimental counts and control were filtered out. The test of significance of ratios was performed by averaging the diploid control and producing a ratio with each biological replicate of the experimental values. Each biological replicate of the treatment was used to produce ratios with the mean of the control. These two sets of log-transformed ratios with base 2 were then compared in the t test. The log of the ratio with base 2 of treatment to the control was plotted on the x axis, while the sum of normalized counts of the treatment and control group was plotted on the y axis. Data points with a P value <0.05 and a corresponding log ratio >0 were depicted in red, while points with a P value <0.05 and a corresponding log ratio <0 were depicted in green. Otherwise they were designated in black.
Gene lists used for the generation of ratio distributions and scatter plots were collected from various resources. For Fig. 1 and SI Appendix, Fig. S2, ratios of all genes were plotted by mapping mRNA reads to the Arabidopsis reference genome TAIR10. For SI Appendix, Fig. S4, the mean of ERCC-normalized counts from diploid plants was computed to determine the relative expression level of genes. We defined genes with ERCC-normalized counts <1 as lowly expressed genes, those with ERCC-normalized counts within the range of 1 to 100 as moderately expressed genes, and those with counts >100 as highly expressed genes (91). For Fig. 2 and SI Appendix, Fig. S5, the list of transcription factors was obtained from PlantTFDB 4.0 (92). Lists of stress-related, signal transduction, proteasome, and ribosomal genes were retrieved from the Arabidopsis reference genome TAIR10 based on their gene annotation. The lists of chloroplast-, mitochondrial, and peroxisomal targeted genes were derived from a published study (93). Determination of mCG(gbM), mCHG, or mCHH status was from the methylation analysis in the present study. The list of genes associated with Polycomb in Arabidopsis was from a published study (51). The list of transcription factors and their targets was from AtTFDB (52, 92). Microarray data from Torres et al. (55) were used for the yeast analysis, and data from Williams et al. (56) were used for the murine analysis.
Transcription Factor Regulation Network Analysis.
The interaction reference data between transcription factors and their target genes were downloaded from AtRegNet, which contains 6,157 predicted and experimentally verified interactions (activation/repression) (52). For each trisomy, we generated a list of genes whose ratios are significantly altered compared with the diploid (generated by edgeR for differential gene expression analysis, P value <0.05) from our dataset (SI Appendix, Table S10). Ratio threshold of transcription factors and target genes in cis is defined as 1.5, whereas that of genes in trans is defined as 1.00. Using the list of statistically significant genes, the following categories were partitioned. If the ratio of a transcription factor is less than the ratio threshold (1.5 or 1, accordingly), and that of its target gene is more than the threshold, then the relationship is defined as “repression.” By contrast, if the ratio of its target gene is less than the threshold, then the relationship is defined as “activation.” Additionally, if the ratio of a transcription factor is over the threshold and that of its target gene is also over the threshold, we define the relationship as activation. If the ratio of the target gene is less than the threshold, the relationship is defined as repression. In this way, the relationship between transcription factors and their target genes is noted by their known relationship in the AtRegNet database (reference activation/reference repression) and from the analyses of our data (data activation/data repression).
Gene Ontology Enrichment Analysis.
Gene Ontology enrichment analysis was performed using the PANTHER Overrepresentation Test tool via Fisher’s exact test with FDR multiple-test correction (94). Categories with FDR <0.05 were defined as statistically significant and are displayed in SI Appendix, Table S4.
Data and Material Availability.
All sequencing data were deposited at the Gene Expression Omnibus (GEO) repository under the accession number GSE79676 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=wpqjcackvdaxvcz&acc=GSE79676). Seeds of the diploid and tetraploid fluorescence-tagged lines are deposited in the Arabidopsis Biological Resource Center, Ohio State University.
Supplementary Material
Acknowledgments
We thank Wen-Ching Chan and Yih-Shien Chiang for the preliminary data analysis and GEO submission. M.M. and A.J.M.M. acknowledge financial support from Academia Sinica and the Taiwan Ministry of Science and Technology (Grants NSC 102-2311-B001-031 and MOST 103-2311-B-001-017). P.-Y.C. is grateful to Academia Sinica and the Taiwan Ministry of Science and Technology for financial support (Projects MOST 103-2313-B-001-003-MY3 and 104-2923-B-001-003-MY2). The University of Missouri group is supported by NSF Grant IOS-1545780. T.J. is supported by NSF Grant Award 1615789.
Footnotes
The authors declare no conflict of interest.
Data deposition: Sequencing data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?token=wpqjcackvdaxvcz&acc=GSE79676 (accession no. GSE79676).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807796115/-/DCSupplemental.
References
- 1.Blakeslee AF, Belling J, Farnham ME. Chromosomal duplication and Mendelian phenomena in Datura mutants. Science. 1920;52:388–390. doi: 10.1126/science.52.1347.388. [DOI] [PubMed] [Google Scholar]
- 2.Blakeslee AF. New Jimson weeds from old chromosomes. J Hered. 1934;25:81–108. [Google Scholar]
- 3.Bridges CB. Sex in relation to chromosomes and genes. Am Nat. 1925;59:127–137. [Google Scholar]
- 4.Birchler JA. A study of enzyme activities in a dosage series of the long arm of chromosome one in maize. Genetics. 1979;92:1211–1229. doi: 10.1093/genetics/92.4.1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Birchler JA, Newton KJ. Modulation of protein levels in chromosomal dosage series of maize: The biochemical basis of aneuploid syndromes. Genetics. 1981;99:247–266. doi: 10.1093/genetics/99.2.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guo M, Birchler JA. Trans-acting dosage effects on the expression of model gene systems in maize aneuploids. Science. 1994;266:1999–2002. doi: 10.1126/science.266.5193.1999. [DOI] [PubMed] [Google Scholar]
- 7.Sun L, et al. Dosage compensation and inverse effects in triple X metafemales of Drosophila. Proc Natl Acad Sci USA. 2013;110:7383–7388. doi: 10.1073/pnas.1305638110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sun L, et al. Differential effect of aneuploidy on the X chromosome and genes with sex-biased expression in Drosophila. Proc Natl Acad Sci USA. 2013;110:16514–16519. doi: 10.1073/pnas.1316041110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sheltzer JM, Torres EM, Dunham MJ, Amon A. Transcriptional consequences of aneuploidy. Proc Natl Acad Sci USA. 2012;109:12644–12649. doi: 10.1073/pnas.1209227109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dürrbaum M, Storchová Z. Effects of aneuploidy on gene expression: Implications for cancer. FEBS J. 2016;283:791–802. doi: 10.1111/febs.13591. [DOI] [PubMed] [Google Scholar]
- 11.Birchler JA, Bhadra U, Bhadra MP, Auger DL. Dosage-dependent gene regulation in multicellular eukaryotes: Implications for dosage compensation, aneuploid syndromes, and quantitative traits. Dev Biol. 2001;234:275–288. doi: 10.1006/dbio.2001.0262. [DOI] [PubMed] [Google Scholar]
- 12.Seidman JG, Seidman C. Transcription factor haploinsufficiency: When half a loaf is not enough. J Clin Invest. 2002;109:451–455. doi: 10.1172/JCI15043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kondrashov FA, Koonin EV. A common framework for understanding the origin of genetic dominance and evolutionary fates of gene duplications. Trends Genet. 2004;20:287–290. doi: 10.1016/j.tig.2004.05.001. [DOI] [PubMed] [Google Scholar]
- 14.Boell L, et al. Exploring the effects of gene dosage on mandible shape in mice as a model for studying the genetic basis of natural variation. Dev Genes Evol. 2013;223:279–287. doi: 10.1007/s00427-013-0443-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Stranger BE, et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ionita-Laza I, Rogers AJ, Lange C, Raby BA, Lee C. Genetic association analysis of copy-number variation (CNV) in human disease pathogenesis. Genomics. 2009;93:22–26. doi: 10.1016/j.ygeno.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Makino T, McLysaght A. Ohnologs in the human genome are dosage balanced and frequently associated with disease. Proc Natl Acad Sci USA. 2010;107:9270–9274. doi: 10.1073/pnas.0914697107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Rice AM, McLysaght A. Dosage sensitivity is a major determinant of human copy number variant pathogenicity. Nat Commun. 2017;8:14366. doi: 10.1038/ncomms14366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wolfe KH, Shields DC. Molecular evidence for an ancient duplication of the entire yeast genome. Nature. 1997;387:708–713. doi: 10.1038/42711. [DOI] [PubMed] [Google Scholar]
- 20.Papp B, Pál C, Hurst LD. Dosage sensitivity and the evolution of gene families in yeast. Nature. 2003;424:194–197. doi: 10.1038/nature01771. [DOI] [PubMed] [Google Scholar]
- 21.Blanc G, Wolfe KH. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–1691. doi: 10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Maere S, et al. Modeling gene and genome duplications in eukaryotes. Proc Natl Acad Sci USA. 2005;102:5454–5459. doi: 10.1073/pnas.0501102102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thomas BC, Pedersen B, Freeling M. Following tetraploidy in an Arabidopsis ancestor, genes were removed preferentially from one homeolog leaving clusters enriched in dose-sensitive genes. Genome Res. 2006;16:934–946. doi: 10.1101/gr.4708406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Simillion C, Vandepoele K, Van Montagu MC, Zabeau M, Van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc Natl Acad Sci USA. 2002;99:13627–13632. doi: 10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Blomme T, et al. The gain and loss of genes during 600 million years of vertebrate evolution. Genome Biol. 2006;7:R43. doi: 10.1186/gb-2006-7-5-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Aury J-M, et al. Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature. 2006;444:171–178. doi: 10.1038/nature05230. [DOI] [PubMed] [Google Scholar]
- 27.Freeling M, et al. Many or most genes in Arabidopsis transposed after the origin of the order Brassicales. Genome Res. 2008;18:1924–1937. doi: 10.1101/gr.081026.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tasdighian S, et al. Reciprocally retained genes in the angiosperm lineage show the hallmarks of dosage balance sensitivity. Plant Cell. 2017;29:2766–2785. doi: 10.1105/tpc.17.00313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Freeling M. Bias in plant gene content following different sorts of duplication: Tandem, whole-genome, segmental, or by transposition. Annu Rev Plant Biol. 2009;60:433–453. doi: 10.1146/annurev.arplant.043008.092122. [DOI] [PubMed] [Google Scholar]
- 30.Emery M, et al. Preferential retention of genes from one parental genome after polyploidy illustrates the nature and scope of the genomic conflicts induced by hybridization. PLoS Genet. 2018;14:e1007267. doi: 10.1371/journal.pgen.1007267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lovén J, et al. Revisiting global gene expression analysis. Cell. 2012;151:476–482. doi: 10.1016/j.cell.2012.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Birchler JA. The genetic basis of dosage compensation of alcohol dehydrogenase-1 in maize. Genetics. 1981;97:625–637. doi: 10.1093/genetics/97.3-4.625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller M, Zhang C, Chen ZJ. Ploidy and hybridity effects on growth vigor and gene expression in Arabidopsis thaliana hybrids and their parents. G3 (Bethesda) 2012;2:505–513. doi: 10.1534/g3.112.002162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Tsukaya H. Does ploidy level directly control cell size? Counterevidence from Arabidopsis genetics. PLoS One. 2013;8:e83729. doi: 10.1371/journal.pone.0083729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Birchler JA, Veitia RA. Gene balance hypothesis: Connecting issues of dosage sensitivity across biological disciplines. Proc Natl Acad Sci USA. 2012;109:14746–14753. doi: 10.1073/pnas.1207726109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Henry IM, Dilkes BP, Miller ES, Burkart-Waco D, Comai L. Phenotypic consequences of aneuploidy in Arabidopsis thaliana. Genetics. 2010;186:1231–1245. doi: 10.1534/genetics.110.121079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Casanova-Sáez R, Candela H, Micol JL. Combined haploinsufficiency and purifying selection drive retention of RPL36a paralogs in Arabidopsis. Sci Rep. 2014;4:4122. doi: 10.1038/srep04122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohashi A, et al. Aneuploidy generates proteotoxic stress and DNA damage concurrently with p53-mediated post-mitotic apoptosis in SAC-impaired cells. Nat Commun. 2015;6:7668. doi: 10.1038/ncomms8668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Coate JE, Schlueter JA, Whaley AM, Doyle JJ. Comparative evolution of photosynthetic genes in response to polyploid and nonpolyploid duplication. Plant Physiol. 2011;155:2081–2095. doi: 10.1104/pp.110.169599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kerkel K, et al. Altered DNA methylation in leukocytes with trisomy 21. PLoS Genet. 2010;6:e1001212. doi: 10.1371/journal.pgen.1001212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckmann-Scholz C, et al. DNA-methylation profiling of fetal tissues reveals marked epigenetic differences between chorionic and amniotic samples. PLoS One. 2012;7:e39014. doi: 10.1371/journal.pone.0039014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jones MJ, et al. Distinct DNA methylation patterns of cognitive impairment and trisomy 21 in Down syndrome. BMC Med Genomics. 2013;6:58. doi: 10.1186/1755-8794-6-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bacalini MG, et al. Identification of a DNA methylation signature in blood cells from persons with Down syndrome. Aging (Albany NY) 2015;7:82–96. doi: 10.18632/aging.100715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mendioroz M, et al. Trans effects of chromosome aneuploidies on DNA methylation patterns in human Down syndrome and mouse models. Genome Biol. 2015;16:263. doi: 10.1186/s13059-015-0827-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sailani MR, et al. DNA methylation patterns in trisomy 21 using cells from monozygotic twins. PLoS One. 2015;10:e0135555. doi: 10.1371/journal.pone.0135555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hatt L, et al. Microarray-based analysis of methylation of 1st trimester trisomic placenta from Down syndrome, Edwards syndrome and Patau syndrome. PLoS One. 2016;11:e0160319. doi: 10.1371/journal.pone.0160319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lim JH, et al. Comprehensive investigation of DNA methylation and gene expression in trisomy 21 placenta. Placenta. 2016;42:17–24. doi: 10.1016/j.placenta.2016.03.012. [DOI] [PubMed] [Google Scholar]
- 48.Alves da Silva AF, et al. Trisomy 21 alters DNA methylation in parent-of-origin-dependent and -independent manners. PLoS One. 2016;11:e0154108. doi: 10.1371/journal.pone.0154108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Niederhuth CE, et al. Widespread natural variation of DNA methylation within angiosperms. Genome Biol. 2016;17:194. doi: 10.1186/s13059-016-1059-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stroud H, et al. Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol. 2014;21:64–72. doi: 10.1038/nsmb.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deleris A, et al. Loss of the DNA methyltransferase MET1 induces H3K9 hypermethylation at PcG target genes and redistribution of H3K27 trimethylation to transposons in Arabidopsis thaliana. PLoS Genet. 2012;8:e1003062. doi: 10.1371/journal.pgen.1003062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Palaniswamy SK, et al. AGRIS and AtRegNet. A platform to link cis-regulatory elements and transcription factors into regulatory networks. Plant Physiol. 2006;140:818–829. doi: 10.1104/pp.105.072280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.McCarthy DJ, Chen Y, Smyth GK. Differential expression analysis of multifactor RNA-seq experiments with respect to biological variation. Nucleic Acids Res. 2012;40:4288–4297. doi: 10.1093/nar/gks042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Torres EM, et al. Effects of aneuploidy on cellular physiology and cell division in haploid yeast. Science. 2007;317:916–924. doi: 10.1126/science.1142210. [DOI] [PubMed] [Google Scholar]
- 56.Williams BR, et al. Aneuploidy affects proliferation and spontaneous immortalization in mammalian cells. Science. 2008;322:703–709. doi: 10.1126/science.1160058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Birchler JA. Facts and artifacts in studies of gene expression in aneuploids and sex chromosomes. Chromosoma. 2014;123:459–469. doi: 10.1007/s00412-014-0478-5. [DOI] [PubMed] [Google Scholar]
- 58.Sinnott EW, Blakeslee AF. Structural changes associated with factor mutations and with chromosome mutations in Datura. Proc Natl Acad Sci USA. 1922;8:17–19. doi: 10.1073/pnas.8.2.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Steinitz-Sears LM. Chromosome studies in Arabidopsis thaliana. Genetics. 1963;48:483–490. doi: 10.1093/genetics/48.4.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Robinson DO, et al. Ploidy and size at multiple scales in the Arabidopsis sepal. Plant Cell. August 24, 2018 doi: 10.1105/tpc.18.00344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Sinnott EW, Houghtaling H, Blakeslee AF. 1934. The Comparative Anatomy of Extra-Chromosomal Types in Datura stramonium (Carnegie Institution of Washington, Washington, DC)
- 62.Lo K-L, et al. Transcriptional consequence and impaired gametogenesis with high-grade aneuploidy in Arabidopsis thaliana. PLoS One. 2014;9:e114617. doi: 10.1371/journal.pone.0114617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Edger PP, et al. Subgenome dominance in an interspecific hybrid, synthetic allopolyploid, and a 140-year-old naturally established neo-allopolyploid monkeyflower. Plant Cell. 2017;29:2150–2167. doi: 10.1105/tpc.17.00010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Muller HJ. Evidence of the precision of genetic adaptation. Harvey Lect. 1950;43:165–229. [Google Scholar]
- 65.Bray D, Lay S. Computer-based analysis of the binding steps in protein complex formation. Proc Natl Acad Sci USA. 1997;94:13493–13498. doi: 10.1073/pnas.94.25.13493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Veitia RA. Exploring the etiology of haploinsufficiency. BioEssays. 2002;24:175–184. doi: 10.1002/bies.10023. [DOI] [PubMed] [Google Scholar]
- 67.Veitia RA. Nonlinear effects in macromolecular assembly and dosage sensitivity. J Theor Biol. 2003;220:19–25. doi: 10.1006/jtbi.2003.3105. [DOI] [PubMed] [Google Scholar]
- 68.Birchler JA, Riddle NC, Auger DL, Veitia RA. Dosage balance in gene regulation: Biological implications. Trends Genet. 2005;21:219–226. doi: 10.1016/j.tig.2005.02.010. [DOI] [PubMed] [Google Scholar]
- 69.Veitia RA, Bottani S, Birchler JA. Cellular reactions to gene dosage imbalance: Genomic, transcriptomic and proteomic effects. Trends Genet. 2008;24:390–397. doi: 10.1016/j.tig.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 70.Walley JW, et al. Integration of omic networks in a developmental atlas of maize. Science. 2016;353:814–818. doi: 10.1126/science.aag1125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: From transcript synthesis to protein degradation. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vavouri T, Semple JI, Garcia-Verdugo R, Lehner B. Intrinsic protein disorder and interaction promiscuity are widely associated with dosage sensitivity. Cell. 2009;138:198–208. doi: 10.1016/j.cell.2009.04.029. [DOI] [PubMed] [Google Scholar]
- 73.Devlin RH, Holm DG, Grigliatti TA. The influence of whole-arm trisomy on gene expression in Drosophila. Genetics. 1988;118:87–101. doi: 10.1093/genetics/118.1.87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Malone JH, et al. Mediation of Drosophila autosomal dosage effects and compensation by network interactions. Genome Biol. 2012;13:r28. doi: 10.1186/gb-2012-13-4-r28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lee H, et al. Effects of gene dose, chromatin, and network topology on expression in Drosophila melanogaster. PLoS Genet. 2016;12:e1006295. doi: 10.1371/journal.pgen.1006295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li Z, et al. Multiple large-scale gene and genome duplications during the evolution of hexapods. Proc Natl Acad Sci USA. 2018;115:4713–4718. doi: 10.1073/pnas.1710791115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sears ER. Cytogenetic studies with polyploid species of wheat. II. Additional chromosomal aberrations in Triticum vulgare. Genetics. 1944;29:232–246. doi: 10.1093/genetics/29.3.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Altug-Teber O, et al. Specific transcriptional changes in human fetuses with autosomal trisomies. Cytogenet Genome Res. 2007;119:171–184. doi: 10.1159/000112058. [DOI] [PubMed] [Google Scholar]
- 79.Do C, Xing Z, Yu YE, Tycko B. Trans-acting epigenetic effects of chromosomal aneuploidies: Lessons from Down syndrome and mouse models. Epigenomics. 2017;9:189–207. doi: 10.2217/epi-2016-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Raznahan A, et al. Sex-chromosome dosage effects on gene expression in humans. Proc Natl Acad Sci USA. 2018;115:7398–7403. doi: 10.1073/pnas.1802889115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Bellott DW, et al. Avian W and mammalian Y chromosomes convergently retained dosage-sensitive regulators. Nat Genet. 2017;49:387–394. doi: 10.1038/ng.3778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bellott DW, et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature. 2014;508:494–499, and erratum (2014) 514:126. doi: 10.1038/nature13206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Pessia E, Makino T, Bailly-Bechet M, McLysaght A, Marais GAB. Mammalian X chromosome inactivation evolved as a dosage-compensation mechanism for dosage-sensitive genes on the X chromosome. Proc Natl Acad Sci USA. 2012;109:5346–5351. doi: 10.1073/pnas.1116763109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Xu J, et al. Free-living human cells reconfigure their chromosomes in the evolution back to uni-cellularity. eLife. 2017;6:e28070. doi: 10.7554/eLife.28070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Matzke AJM, Watanabe K, van der Winden J, Naumann U, Matzke M. High frequency, cell type-specific visualization of fluorescent-tagged genomic sites in interphase and mitotic cells of living Arabidopsis plants. Plant Methods. 2010;6:2. doi: 10.1186/1746-4811-6-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Matzke AJM, Huettel B, van der Winden J, Matzke M. Use of two-color fluorescence-tagged transgenes to study interphase chromosomes in living plants. Plant Physiol. 2005;139:1586–1596. doi: 10.1104/pp.105.071068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Huettel B, Kreil DP, Matzke M, Matzke AJM. Effects of aneuploidy on genome structure, expression, and interphase organization in Arabidopsis thaliana. PLoS Genet. 2008;4:e1000226. doi: 10.1371/journal.pgen.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kim D, et al. TopHat2: Accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Trapnell C, et al. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wickham H. ggplot2: Elegant Graphics for Data Analysis. Springer; New York: 2009. [Google Scholar]
- 91.Jiang H, Wong WH. Statistical inferences for isoform expression in RNA-seq. Bioinformatics. 2009;25:1026–1032. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Jin J, et al. PlantTFDB 4.0: Toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2017;45:D1040–D1045. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Law SR, et al. Nucleotide and RNA metabolism prime translational initiation in the earliest events of mitochondrial biogenesis during Arabidopsis germination. Plant Physiol. 2012;158:1610–1627. doi: 10.1104/pp.111.192351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Thomas PD, et al. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.