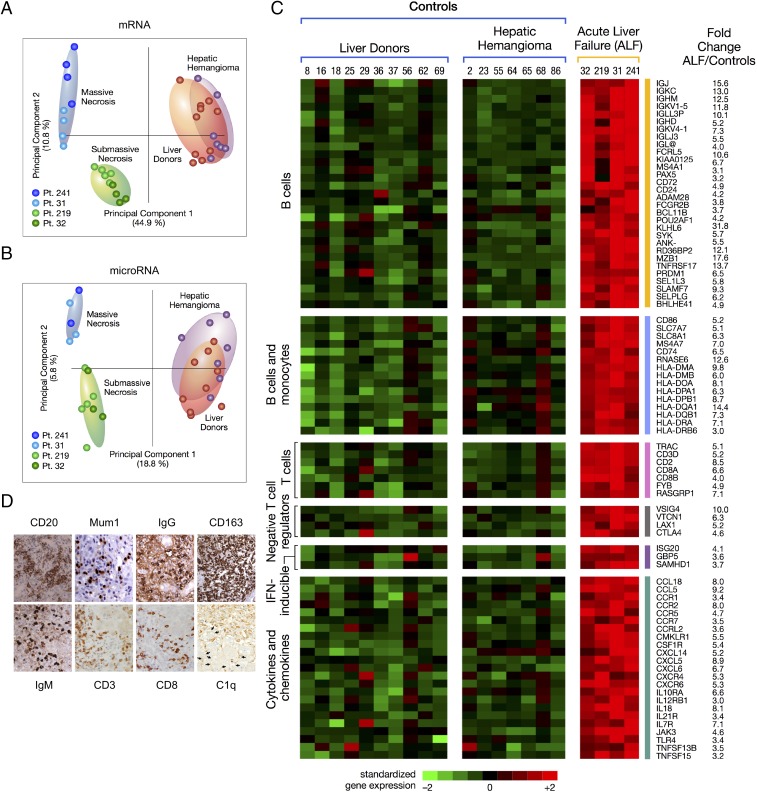
Fig. 5.
Principal component analysis of mRNA and miRNA, heat map of differentially expressed immune genes, and immunohistochemistry in the liver of patients with HBV-associated ALF. (A) Unsupervised principal component analyses of 6,996 mRNAs and (B) 1,105 mature miRNAs from four patients with ALF. The analysis also includes 17 samples from control livers, including 10 liver donors and 7 subjects who underwent liver resection for hepatic hemangioma. Each dot represents a single liver sample. Both plots show a complete separation between ALF and control livers. Within the ALF group, a separation was also observed between massive and submassive hepatic necrosis. (C) Heat map of immune genes differentially expressed in ALF. Each column represents a single patient. Data from the four ALF patients represent the average of multiple specimens analyzed (up to five liver specimens for each patient), whereas data from the 17 controls represent individual liver samples. Each cell represents the expression of a particular gene (rows) of a particular liver specimen (columns). Gene expression levels were log2-transformed and rowwise-standardized. Up-regulated genes are shown in shades of red and down-regulated genes in shades of green. (D) Immunohistochemical staining of liver tissue in ALF confirmed the prominent B cell signature with extensive infiltration of B cells (CD20), plasma blasts and plasma cells (Mum1), with strong cytoplasmic staining for IgM and IgG antibodies. The liver sections also showed the presence of T cells (CD3), mostly CD8-positive, extensive macrophage infiltration (CD163), and deposition of complement factor C1q. The arrows indicate weak linear intercellular staining of residual hepatocytes in a patient with submassive hepatic necrosis (magnification 40×).