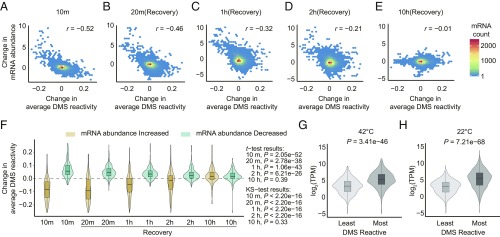
Fig. 4.
Strong negative correlation between heat-shock-induced DMS reactivity change and heat-shock-induced mRNA abundance (TPM) change that gradually dissipates after heat shock. (A–E) Change of average DMS reactivity (42 °C − 22 °C) from Structure-seq (all 10 min) vs. fold change (log2) in mRNA abundance (42 °C − 22 °C) from RNA-seq (see Fig. 1A for time course), calculated on all mRNAs with sufficient Structure-seq coverage. (A) 10 min (= end of 42 °C treatment), (B) 20 min, (C) 1 h, (D) 2 h, (E) 10 h. (F) Distribution of change in average DMS reactivity of all transcripts with sufficient Structure-seq coverage within the top 5% of mRNAs with increased abundance (beige) and the bottom 5% of mRNAs with decreased abundance (blue). (G and H) The abundance of degradome fragments of the top/bottom 5% most/least DMS reactive transcripts at (G) 42 °C and (H) 22 °C is compared, showing that more reactive transcripts have a higher mean number of degradome fragments.