Significance
All known naturally occurring red fluorescent proteins (RFPs), a class that is desirable for biological imaging, are tetrameric, limiting their usefulness as molecular fusion tags in in vivo model systems. Here we explore protein variant libraries targeted at monomerizing far-red RFP variants and describe a generalizable method to monomerize RFPs of interest. This method preserves the fluorescence of the molecule throughout its monomerization, in contrast to break–fix methods, allowing selective enrichment of bright, far-red monomeric variants. Furthermore, we report four bright monomeric RFPs here, which are among the most red-shifted of any monomeric Aequorea victoria-class FPs.
Keywords: fluorescent protein, red fluorescent protein, protein engineering, computational protein design, RFP
Abstract
Anthozoa-class red fluorescent proteins (RFPs) are frequently used as biological markers, with far-red (λem ∼ 600–700 nm) emitting variants sought for whole-animal imaging because biological tissues are more permeable to light in this range. A barrier to the use of naturally occurring RFP variants as molecular markers is that all are tetrameric, which is not ideal for cell biological applications. Efforts to engineer monomeric RFPs have typically produced dimmer and blue-shifted variants because the chromophore is sensitive to small structural perturbations. In fact, despite much effort, only four native RFPs have been successfully monomerized, leaving the majority of RFP biodiversity untapped in biomarker development. Here we report the generation of monomeric variants of HcRed and mCardinal, both far-red dimers, and describe a comprehensive methodology for the monomerization of red-shifted oligomeric RFPs. Among the resultant variants is mKelly1 (emission maximum, λem = 656 nm), which, along with the recently reported mGarnet2 [Matela G, et al. (2017) Chem Commun (Camb) 53:979–982], forms a class of bright, monomeric, far-red FPs.
The development of red fluorescent proteins (RFPs) as tags for molecular imaging has long focused on monomerization, increased brightness, and pushing excitation and emission to ever-longer wavelengths. These traits are desirable for live animal imaging because far-red and near-infrared light penetrates tissue with minimal absorption in what is known as the near infrared window (∼625–1,300 nm) (1, 2). Monomericity is important because oligomerization of a fluorescent protein (FP) tag can artificially aggregate its linked protein target, altering diffusion rates and interfering with target transport, trafficking, and activity (3, 4). Recently, a new class of infrared fluorescent proteins (iRFPs) was developed from the bacterial phytochrome (5, 6), but these require the covalent linkage of a small molecule chromophore, biliverdin, limiting their use to cells and organisms that make this molecule in sufficient quantity. Anthozoa-class RFPs (such as mCherry and mKate) have the advantage that the chromophore is created via a self-catalyzed reaction, necessitating only molecular O2 for chromophore formation (7), and have been engineered to exhibit peak fluorescence at wavelengths as long as 675–685 nm (8, 9).
To our knowledge, ∼50 native RFPs and ∼40 chromoproteins (CPs) with peak absorbance in the red or far-red (absorbance maximum, λabs > 550 nm) have been described to date, but most have not been extensively characterized because they are as a class tetrameric and thus are less useful as biological markers (10, 11). An underlying biological reason for the obligate tetramerization of native RFPs has been suggested but is not well understood (12–15). Oligomerization does seem to play an important structural role, however, because breaking tetramerization without abrogating fluorescence has proved difficult, and successful monomerization has always led to either a hypsochromic shift in λem or a decrease in brightness (16–19). Previous efforts to monomerize native RFP tetramers have relied on lengthy engineering trajectories, with only four native RFPs having been successfully monomerized before this work (Table 1). Generally, mutations are first introduced into protein/protein interfaces to weaken oligomerization, an inefficient process that compromises fluorescence, and then random mutagenesis and screening are used to isolate variants with partially recovered fluorescence. After many such cycles, monomeric variants have been found, but protein core and chromophore-proximal mutations are invariably introduced, making it difficult to exert control over the fluorescent properties of the resultant monomer. It is thus difficult to know whether the poor spectroscopic characteristics of engineered monomers are an unavoidable consequence of monomerization or only the manifestation of a suboptimal evolutionary path. The engineering of mScarlet, a bright red monomer that was designed synthetically from previous RFP monomers, lends evidence in support of the poor characteristics of monomers not being intrinsic to the monomeric scaffold (20).
Table 1.
Engineered monomeric RFPs
| Monomeric RFP | Brightness (Φ × ε)/1,000 | λem, nm | Mutations to core | Total mutations | Immediate parent (dimer/tetramer) | Brightness (Φ × ε)/1,000 | λem, nm | Mutations to core | Total mutations | Ancestral parent (dimer/tetramer) | Brightness (Φ × ε)/1,000 | λem, nm |
| mRFP1 | 12.5 | 607 | 13 | 33 | – | – | – | – | – | DsRed (T) | 59.3 | 583 |
| DsRed.M1 | 3.5 | 586 | 10 | 45 | – | – | – | – | – | DsRed (T) | 59.3 | 583 |
| FusionRed | 18.0 | 608 | 9 | 45 | mKate2 (D) | 18.0 | 630 | 7 | 27 | eqFP578 (T) | 55.1 | 578 |
| mRuby | 39.2 | 605 | 6 | 40 | – | – | – | – | – | eqFP611 (T) | 35.1 | 611 |
| mKeima | 3.5 | 620 | 7 | 17 | dKeima (D) | 7.6 | 616 | 5 | 13 | COCP (T) | n/a | n/a |
| mGinger1* | 1.2 | 637 | 7 | 45 | HcRed7 (D) | 6.0 | 645 | 4 | 8 | hcriCP (T) | n/a | n/a |
| mGinger2* | 1.4 | 631 | 7 | 49 | HcRed7 (D) | 6.0 | 645 | 4 | 8 | hcriCP (T) | n/a | n/a |
| mKelly1* | 7.0 | 656 | 15 | 52 | mCardinal (D) | 12.8 | 656 | 15 | 44 | eqFP578 (T) | 55.1 | 578 |
| mKelly2* | 7.7 | 649 | 15 | 52 | mCardinal (D) | 12.8 | 656 | 15 | 44 | eqFP578 (T) | 55.1 | 578 |
All known native RFPs are tetrameric. A common trajectory in the engineering of a monomeric derivative of a tetrameric RFP first passes through a dimeric intermediate. Dashes represent instances for which there is no intermediate parent protein. n/a, not applicable.
This work.
Here we present a comprehensive engineering strategy for the monomerization of RFPs that differentiates itself by treating separately the problems of protein stabilization, core optimization, and surface design. We sample mutational space both stochastically, through error-prone PCR (ePCR) mutagenesis, and rationally, by analysis of multiple sequence alignments (MSAs) and computational protein design (CPD). Two far-red oligomeric proteins were targeted for monomerization: HcRed (λem = 633 nm), a dimer/tetramer (21), and mCardinal (λem = 656 nm), a reported monomer that in our hands is dimeric. The monomeric RFPs reported here include two monomeric HcRed variants, mGinger1 (λem = 637 nm) and mGinger2 (λem = 631), and two monomeric mCardinal variants, mKelly1 (λem = 656 nm) and mKelly2 (λem = 649 nm), which are among the brightest far-red monomeric FPs to have been reported.
Results
Stepwise Monomerization of HcRed.
We first chose HcRed, a far-red FP that has been engineered but never successfully monomerized (21, 22). As we have previously demonstrated, oligomericity and brightness can be treated as separate protein design problems (23). We devised a workflow that separately targets the chromophore environment (to engineer a protein core that maintains structural integrity absent stabilizing oligomeric interactions) and the protein surface (to drive monomerization). Anthozoa-class RFPs have two oligomeric interfaces, named AB and AC (24), with the AC interface being the more stable of the two and burying a large hydrophobic surface (25). Early engineering of HcRed partially disrupted oligomerization at the AB interface, but all mutations to the AC interface were found to vitiate fluorescence (21). To test the integrity of the AC interface, we made successive deletions to HcRed’s C-terminal tail (residues 219–227), which plays an integral role in the AC interaction (Fig. 1A). HcRed lost significant brightness with the deletion of just one C-terminal residue, N227, and was nonfluorescent after any further deletion, demonstrating that optimization would be necessary before monomerization.
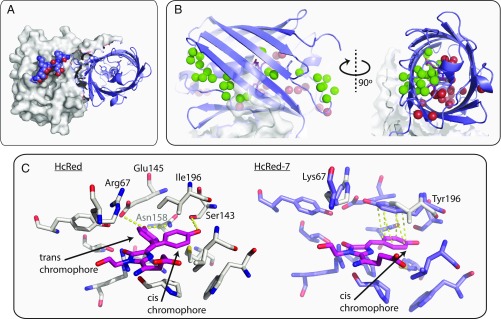
Fig. 1.
The structure of HcRed7 (PDB ID 6DEJ), a far-red dimer. (A) The HcRed7 dimeric interface is stabilized by its C-terminal tail. One monomer is shown as a cartoon, while the second is shown as a surface; residues 222–227 are shown in spheres. (B) A water channel stretches from a gap in HcRed7’s β-barrel through the center of the protein to a smaller opening at its outlet on the other end of the barrel. (C, Left) The crystal structure of HcRed (PDB ID: 1YZW; in gray sticks) showing dual occupancy of the chromophore’s phenolate group. The cis chromophore is stabilized by a C143S mutation from parent protein hcriCP. The trans chromophore is stabilized by two hydrogen bonds (in yellow) from Glu145 and Asn158. (C, Right) Two mutations (R67K and I196Y) were made in the HcRed background to create HcRed7 (slate sticks). Tyr196 in HcRed7 stabilizes the cis chromophore with a π-stacking interaction (in yellow).
We endeavored to engineer a more stable core, identifying two mutational hot spots from an alignment of far-red RFPs (SI Appendix, Table S1): (i) a group of residues that surrounds alternative conformations of the chromophore’s phenolate ring and (ii) a region above the plane of the chromophore, between the central α-helix and the AC oligomeric interface (SI Appendix, Fig. S1). Generally in RFPs, the cis chromophore—phenolate ring is cis to the proximal nitrogen on the imidazolinone ring—is the fluorescent species (26). In engineering HcRed from its chromoprotein parent hcriCP, the cis chromophore was stabilized over the nonfluorescent trans chromophore by way of a cysteine to serine mutation at position 143, which provides a hydrogen bond to the cis phenolate oxygen (Fig. 1C) (27). We reasoned that further stabilization of the cis chromophore would increase brightness and designed two libraries using HcRed as the parent. The first core library (cLibA) targeted hot spot A, mutating trans-stabilizing amino acids, placing bulkier side chains into the trans pocket, and allowing varied hydrogen bonding geometries to the cis chromophore. A second core library (cLibB) targeted hot spot B along with two chromophore-backing positions, Gly28 and Met41, that are implicated in maturation and color (8, 25, 28). Two key features of this hot spot are a channel populated by structural water molecules that stretches to the protein surface and Arg67, a key catalytic residue for chromophore formation (27). Mutations to this region may serve to occlude access to the chromophore by bulk solvent upon monomerization and to allow room for chromophore processing. Small libraries of fewer than 1,000 protein variants were guided by the far-red RFP alignment (SI Appendix, Table S1). After screening each library to >95% coverage, we fully characterized 16 cLibA variants and 21 cLibB variants. The variants showed brightness increases of up to 10-fold and displayed a broad range of emission profiles, with λem between 606 and 647 nm (SI Appendix, Fig. S2). To determine which if any variants would be amenable to monomerization, we tested a five-residue tail deletion (amino acids 223–227) to each of the 41 cLibA and cLibB variants. Eight of these variants showed detectable fluorescence after the tail deletion. A double mutant (R67K/I196Y) designated HcRed7 (λem = 645 nm) produced the most red-shifted of the fluorescent tail-deleted variants, HcRed7∆5 (λem = 643 nm). Relative to HcRed, the core mutations in HcRed7 bathochromically shift its emission by 12 nm, improve its quantum yield (Φ) from 0.05 to 0.08 (P < 0.01), and thermostabilize the protein by 6 °C. HcRed7, however, loses significant brightness with the deletion of six tail residues (HcRed7∆6) and becomes 10 °C less thermostable, indicating that the protein is destabilized by disruption to its oligomerization (Table 2).
Table 2.
Photophysical properties of protein variants derived from HcRed and mCardinal
| Protein | Φ | ε, M−1⋅cm−1 × 10−3 | Brightness (Φ × ε) | λex, nm | λem, nm | pKa | Maturation (T0.5 − min) | Photostability (T0.5 − min) | Apparent Tm, °C |
| HcRed | 0.05 | 70 | 3.5 | 585 | 633 | 4.0/10.0 | 59 | 36 | 69 |
| HcRed7 | 0.08 | 75 | 6.0 | 592 | 645 | 75 | |||
| HcRed7∆5 | 0.06 | 69 | 4.1 | 592 | 643 | 71 | |||
| HcRed7∆6 | * | * | 582 | 635 | 65 | ||||
| HcRed77∆6 | 0.05 | † | † | 68 | |||||
| HcRedm1 | 0.01 | † | † | 64 | |||||
| mGinger1 | 0.02 | 58 | 1.2 | 587 | 637 | 7.9 | 106 | 63 | 79 |
| mGinger2 | 0.04 | 36 | 1.4 | 578 | 631 | 6.5 | 74 | 17 | 80 |
| mCardinal | 0.16 | 80 | 12.8 | 601 | 656 | 4.6 | 20 | 26 | |
| mCardinal-mut6∆19 | 0.13 | 60 | 7.8 | 596 | 649 | ||||
| mKelly1 | 0.16 | 44 | 7.0 | 596 | 656 | 5.4 | 28 | 7.0 | |
| mKelly2 | 0.18 | 43 | 7.7 | 598 | 649 | 5.6 | 20 | 5.1 |
Values of λem and λem are the maximum wavelengths of the visible excitation and emission spectra, respectively; Φ is the quantum yield; and ε is the extinction coefficient at the absorbance peak. Details can be found in Materials and Methods.
Too dim/poorly expressed to measure accurately.
Extinction coefficient (and therefore brightness) could not be measured because of multiple chromophore species present.
To further optimize HcRed7∆6 for monomerization, we took aim at improving the thermostability of the protein. Thermostability has been shown to increase a protein’s evolvability (29), and consensus design is one of the best tools for improving thermostability (30). We constructed an MSA that consists of 741 Aequorea victoria-class FPs (Materials and Methods) and then built a library in the HcRed7∆6 background to sample all 105 positions in HcRed containing a nonconsensus amino acid with the consensus amino acid (31) and compared this to a strategy of ePCR mutagenesis. We screened the consensus (∼1.2 mutations per variant) and ePCR (∼1.8 mutations per variant) libraries at 675 nm to allow maximal differentiation between far-red variants whose λem was between 630 and 640 nm and a large population of near-red variants whose emission peaked between 605 and 620 nm but which were often brighter. The consensus library was screened to 40× coverage (∼4,300 clones), and ∼8,600 clones were screened from the ePCR library. Consensus library variants significantly outperformed ePCR library variants (SI Appendix, Fig. S3). We combined seven of the top consensus variants together into a chimeric protein, HcRed77∆6, which recovered much of the quantum yield lost with the tail deletion (Table 2).
Finally, to monomerize HcRed77∆6 we targeted the AC interface with a CPD procedure that we described in previous work (23). We focused on a set of five hydrophobic residues (Val146, Val159, Ile170, Phe191, and Phe193) at the heart of the AC interface that make extensive intermolecular contacts (SI Appendix, Fig. S4) and built a 100,000-member combinatorial library guided by the design. We isolated a first-generation monomer, HcRedm1, and verified it to be monomeric by size exclusion chromatography (SEC) and analytical ultracentrifugation (AUC) (Fig. 2). HcRedm1, however, was dim and expressed poorly (Table 2). We attributed these poor attributes to incomplete thermostabilization of the parent, HcRed77∆6, and subsequently used DNA shuffling to sample mutations from the ePCR library. Consensus mutations had been sampled to generate HcRedm1, while ePCR mutations offered the chance to move into novel sequence space. Specifically, we shuffled HcRedm1 with two HcRedm1 variants containing either 13 or 16 of the best candidate mutations from the ePCR library. This library was screened at 37 °C (instead of 30 °C for the earlier screens) and followed by a final round of ePCR mutagenesis to the top hit from the screen. Two bright variants were isolated with improved brightness and thermostability relative to the parent HcRedm1: mGinger1 and mGinger2 (Table 2 and SI Appendix, Fig. S5).
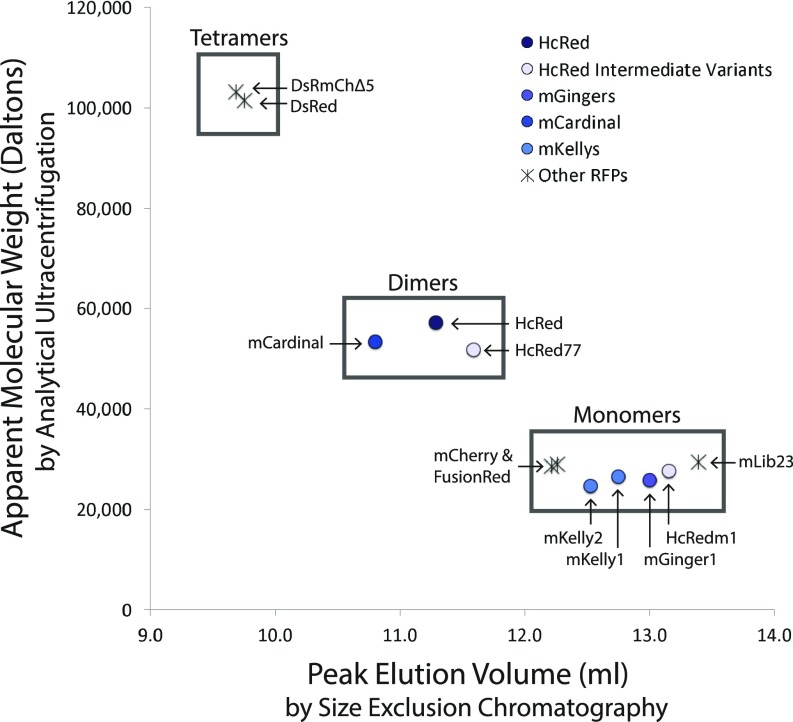
Fig. 2.
Oligomeric analysis of RFPs. The apparent molecular weight as calculated from a c(M) distribution of sedimentation velocity data from an analytical ultracentrifuge run is plotted on the y axis. The x axis shows the peak elution volume as measured at 590 nm absorbance by size exclusion chromatography. Groupings are boxed as monomers, dimers, and tetramers.
Two-Step Monomerization of mCardinal.
The monomerization of HcRed required three design elements, core optimization, protein thermostabilization, and surface design, and utilized mutational diversity from three sources, an MSA, CPD, and ePCR mutagenesis. While the engineering process for the development of the mGingers was rational and therefore involved few rounds of screening, we felt that it could be further improved by integrating the three design objectives into one large library. We targeted mCardinal, a recently reported variant of mNeptune that was reported to be monomeric but that we have shown to be dimeric by both SEC and AUC (Fig. 2). In fact, the crystal structure of mCardinal [Protein Databank (PDB) ID: 4OQW] shows the protein in a classic tetrameric RFP arrangement, similar to DsRed and mCardinal’s progenitor, eqFP578 (32).
As with HcRed, we first probed tail deletion variants of mCardinal, which was previously engineered to have a long, 20-amino acid C-terminal tail (33). The first 15 residues were easily removed (equivalent to HcRed∆4), but as was the case with HcRed, mCardinal∆16 (equivalent to HcRed∆5) is significantly dimmer, and mCardinal∆18 (equivalent to HcRed∆7) is essentially nonfluorescent. To discover mutations for a subsequent combined library approach, we targeted mCardinal∆19, a near-total tail deletion, with ePCR and isolated six mutations that restored measureable fluorescence and did not hypsochromically shift the emission spectrum. The six identified ePCR hits were combined to form mCardinal-mut6∆19, which showed a similar brightness to mCardinal (Table 2). We then built a monomerization library that included the six stabilizing ePCR mutations and a complete tail deletion (∆20) and that sampled a CPD-generated AC interface library and the nine highest-scoring consensus mutations (SI Appendix, Table S2). Because the first-generation HcRed monomer needed further optimization for improved brightness, we chose to sample a larger surface design landscape than we did in the case of HcRed, again designing the five-residue core of the AC interface but also allowing diversity in eight other nearby surface positions (SI Appendix, Table S3). The total theoretical library size was 5.7 × 107. After screening 1.1 × 105 variants by fluorescence-activated cell sorting (FACS), we isolated two variants that were bright and monomeric and retained far-red emission: mKelly1 and mKelly2 (Fig. 2 and Table 2). The monomericity of the mGingers and mKellys was confirmed in live cells using the CytERM assay (Table 3) (34).
Table 3.
Determination of in vivo oligomericity of RFPs by the CytERM assay
| Protein | No. of cells | No. of cells with OSER structures | Percentage of normal cells |
| HcRed | 316 | 313 | 0.9% |
| mGinger1 | 301 | 31 | 89.7% |
| mGinger2 | 273 | 29 | 89.4% |
| mCardinal | 290 | 199 | 31.7% |
| mKelly1 | 274 | 19 | 93.1% |
| mKelly2 | 354 | 107 | 69.8% |
Discussion
Clear Design Objectives Speed Protein Development.
We demonstrate that an engineering process that uses multiple protein engineering approaches can hasten the isolation of optimized protein variants. To develop the mGingers, we utilized multiple rationally guided approaches to design small libraries of diverse but functional HcRed variants, focused separately on the problems of brightness, stability, and oligomericity. Beginning with oligomers partially destabilized by the deletion of HcRed’s C-terminal tail, we explored functional sequence space small libraries and then sampled the combinatorial space of the isolated mutations using DNA shuffling (35, 36), allowing incorporation of 38 and 42 mutations over five rounds of screening into mGinger1 and mGinger2, respectively (Fig. 3 and SI Appendix, Fig. S6). Noting that high-value mutations were enriched during the DNA shuffling-based selection, we streamlined our design procedure, allowing us to monomerize mCardinal in two steps, optimizing first for tail deletion and subsequently screening one large multipurpose combinatorial library for bright monomers. We incorporated 39 and 42 mutations into the resultant monomers, mKelly1 and mKelly2, respectively. Unlike previous RFP monomerization efforts, we maintained fluorescence at every design stage, allowing for screening to maintain the desired far-red emission. The mutations in the final RFP variants were found by employing complementary but divergent engineering processes. Consensus design was used to improve thermostability, which has been shown to improve proteins’ evolvability (29, 30), while ePCR mutagenesis added diversity to this pool of stabilizing mutations. Notably, consensus design significantly outperformed random mutagenesis in improving the brightness of HcRed7 (SI Appendix, Fig. S3). Finally, to build stable and soluble β-sheet surfaces, an application suited to neither consensus design nor ePCR mutagenesis, we used CPD, which we had previously shown to be well suited to this purpose.
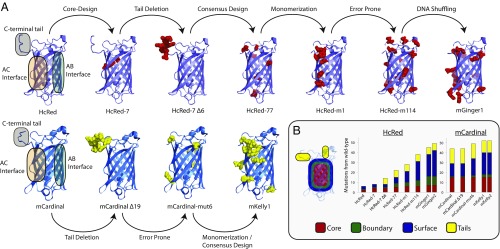
Fig. 3.
Engineering of the mGingers and mKellys. (A) Each engineering step is shown with the mutations made indicated by red spheres in the HcRed background and yellow spheres in the mCardinal background. The crystal structures are of HcRed7 (reported here; PDB ID: 6DEJ) and mCardinal (PDB ID: 4OQW). (B) Solvent-exposed surface area (SASA) was used to categorize the mutations into three buckets, with tails treated separately. The number of mutations separating each designed variant from its wild-type ancestral progenitor (hcriCP in the case of HcRed variants and eqFP578 in the case of mCardinal variants) is plotted.
Mutations Accumulate in Key Structural Regions.
A total of 45 mutations in mGinger1 and 52 mutations in mKelly1 separate them from their progenitors hcriCP and eqFP578, respectively. These mutations cluster structurally: at the designed AC interface, at chromophore-proximal positions, and near pockets of exposed hydrophobic residues on the protein surface. One region of particular note in which mutations cluster is an apparent channel populated by structural water molecules that runs from a 6–13 Å wide cleft in the β-barrel between β-strands 7 and 10, through the chromophore pocket, exiting the other end of the β-barrel near the C-terminal end of the central α-helix and a small 5–8 Å gap between β-strands 3 and 11 (Fig. 1B). These deformations of the β-barrel are bisected by the attachment site of the C-terminal tail and appear to be stabilized by intermolecular interactions between monomers across the AC interface. A break of the AC interface may destabilize the water channel, exposing the chromophore to bulk solvent, which would in turn interfere with chromophore maturation and quench fluorescence (28, 37). Indeed, mGinger1 and mKelly1 have 11 and 6 mutations, respectively, to residues that are in close proximity (4 Å) to structural waters in this channel and that are not a part of the AC interface (SI Appendix, Fig. S7). Elsewhere, mGinger1 and mKelly1 have 11 and 15 mutations, respectively, to their AC interfaces and 2 and 3 mutations, respectively, to their AB interfaces, which likely contribute to breaking oligomerization. In mGinger1 we see eight mutations to patches of exposed hydrophobic surface residues not located at the oligomeric interfaces, as mapped by spatial aggregation propensity (38, 39), whereas with mKelly1, we see only two new surface mutations because we expect that the previous engineering of mCardinal resulted in optimizing its noninterface surfaces for solubility. Outside of these structural clusters, we introduced relatively few new mutations to mGinger1 and mKelly1, five in each case. mKelly1 does inherit 11 other uncharacterized mutations from mCardinal, to both its surface and core.
Protein Stability Is Linked to Function.
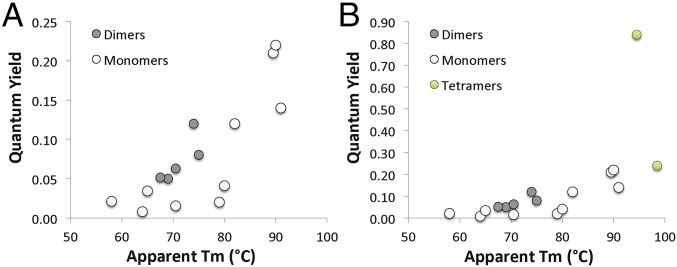
Past efforts to monomerize RFPs have largely ignored the role that scaffold stability may play in engineering a functional monomer. We suggest that as oligomericity is broken, a loss of structural integrity (approximated here by apparent Tm) can leave monomers unstable and nonfunctional. As we monomerized HcRed, we measured the thermal stability of selected intermediates and found a positive correlation between apparent Tm and quantum yield (Fig. 4 and SI Appendix, Table S4). This relationship may be related to scaffold rigidity because in a more rigid excited-state chromophore, there is less nonradiative decay of fluorescent energy via thermal motion or other atomic interactions (40, 41). In small molecule fluorophores this is readily seen, as quantum yield increases with decreased temperature (42). Furthermore, rational design of a chromophore-proximal β-strand was used to improve quantum yield of a cyan FP to 0.93 (40). The correlation between quantum yield and apparent Tm, however, appears to divide into two distinct groups, with dimers having higher quantum yields than monomers. mGinger1 and mGinger2, for instance, despite being thermostabilized by ∼5 °C over the parental protein HcRed7, are less bright. We observe an uncoupling between thermostability and quantum yield, where despite sharing an almost identical protein core, the less thermally stable dimer is brighter than its thermostabilized monomeric derivative. The effect of the oligomericity of the protein scaffold seems to further manifest in that the monomeric variants reported here are less photostable, and in the case of the mGingers also mature more slowly, than the dimeric proteins from which they were derived (Table 2 and SI Appendix, Figs. S8 and S9). The two dimeric proteins studied here show some photoactivation behavior upon illumination (SI Appendix, Fig. S9), similar to that reported to occur because of the stabilization of the cis chromophore via the sulfoxidation of a cysteine residue in a S143C variant of mKate2 (a dimeric RFP) (43). This does not appear to explain the phenomenon in the observed cases in this work, because position 143 is occupied by a threonine in mCardinal and a serine in HcRed (this residue was unmutated during monomerization in both cases).
Fig. 4.
Thermal stability versus quantum yield of HcRed, mCardinal, DsRed, and their variants. (A) Monomers and dimers shown separately for clarity. (B) Monomers, dimers, and tetramers. For a list of proteins included in this figure, see SI Appendix, Table S4.
HcRed7’s Structure Explains Brightness and Bathochromic Emission.
We solved an X-ray crystal structure of HcRed7 (PDB ID: 6DEJ), which shows that the mutation from histidine to tyrosine at position 196 serves to add a π-stacking interaction with the chromophore phenolate ring (Fig. 1C). Tyr196 π-stacks with the fluorescent cis orientation of the phenolate, serving to both stabilize the fluorescent cis phenolate over the nonfluorescent trans phenolate—HcRed’s chromophore occupies both cis and trans conformations—and to red-shift the λem, as a π-stacking phenolate interaction has been shown to reduce the energy of the excited state of the chromophore (44–46). In turn, position 67 is a key catalytic residue that functions as a base, abstracting a proton from the bridging carbon of the phenolate side chain during cyclization (7, 47). This residue is almost invariably a lysine or arginine among RFPs, and we propose that the mutation from arginine to lysine here allows room for the π-stacking interaction and the bulkier tyrosine side chain. It has been previously noted that this π-stacking interaction can induce a bathochromic shift in λem (48), but here we note that these two mutations also conveyed a 6 °C improvement to apparent Tm and a 60% improvement in quantum yield relative to HcRed.
Conclusion
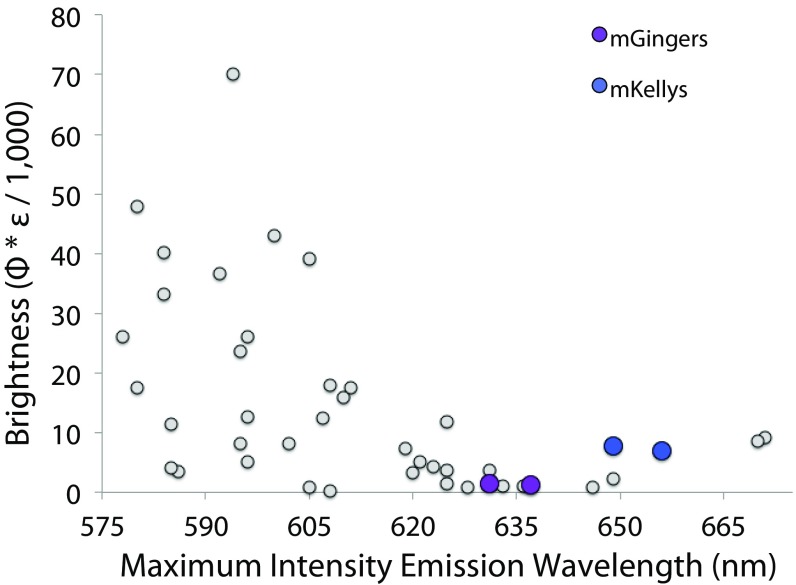
We engineered four monomeric RFPs: mGinger1/2 and mKelly1/2, monomeric variants of the far-red fluorescent proteins HcRed and mCardinal, both dimeric RFPs that had been the targets of previous monomerization attempts. mKelly1 and mKelly2 join mGarnet and mGarnet2 as part of a new class of bright monomeric RFPs with emission peaking near to or longer than 650 nm (Fig. 5 and SI Appendix, Table S5) (49). Previously, we monomerized DsRed using a prestabilized core borrowed from mCherry and showed that monomerization is possible with little to no change to an RFP’s spectroscopic properties (23). Here we show that stabilization of the entire protein scaffold is important for monomerization. Despite the mGingers and mKellys being slightly dimmer and hypsochromically shifted from HcRed7 and mCardinal, they move the needle toward longer wavelengths and brighter emission for monomers. Past monomerization efforts have been beset by similar loss of brightness and hypsochromic shifts, but because they necessitated significant mutation to the core of the protein and the chromophore environment (16–18, 50, 51), it has been difficult to separate the effects of potentially suboptimal core mutation from the effects of monomerization. The rational approach that we lay out in monomerizing HcRed and mCardinal includes elements of rational design, computational design, and directed evolution and represents a marked improvement in the efficiency of RFP monomerization. Further exploration of stable RFP cores will be necessary to determine how to significantly improve brightness postmonomerization.
Fig. 5.
Brightness versus emission maximum for monomer RFPs. For a list of proteins included in this figure, see SI Appendix, Table S5.
Materials and Methods
Plasmids and Bacterial Strains.
The HcRed sequence was taken and modified from the hcriCP GenBank entry (accession no. AF363776). Ten amino acids were added to the N terminus, consisting of a Methionine followed by a 6x Histidine tag for protein purification, followed by a Gly-Ser-Gly linker sequence. All genes were constructed by overlap extension PCR from oligonucleotides designed by DNAworks and ordered from Integrated DNA Technologies (IDT). Assembled genes were PCR-amplified and cloned into the pET-53-DEST expression plasmid (EMD Millipore). Constructs were sequence-verified and transformed into BL21-Gold(DE3) competent cells for protein expression (Agilent).
Construction of Designed Libraries.
The HcRed core variants were designed with DNAworks as mutant runs of the wild-type gene assembly. The HcRed AC surface library used the triplet codon VRN to replace the five design positions, which allows for the possible amino acids D/E/G/H/K/N/Q/R/S. The mCardinal monomer library was designed by hand because it was too complex for DNAworks; we used degenerate bases where possible. For all libraries, oligonucleotides were ordered from IDT, and cloning was carried out as described above.
Error-Prone PCR Mutagenesis.
Error-prone PCR mutagenesis of HcRed variants was performed by addition of manganese chloride to Taq DNA polymerase PCR reactions. Ten, 15, and 20 µM MnCl2 were tested and cloned with PIPE cloning into pET-53-DEST for sequencing. Twelve colonies from each library were picked and sequenced, and the library with a mutation rate closest to but not more than 2.0 mutations per gene was selected for further screening.
DNA Shuffling.
The variants that were to be shuffled together were PCR-amplified and purified by gel electrophoresis with a standard spin-column gel purification kit (Qiagen). Five µg of the purified DNA was then digested with 0.5 U of DNaseI (NEB) in a 50-µL reaction. The reaction was allowed to sit for 7.5 min at room temperature and then quenched with 5 µL of 100 mM EDTA (4× the concentration of MgCl2 in the reaction buffer). The reaction was further heat-inactivated for 10 min at 90 °C in a thermocycler and electrophoresed. Bands of ∼30 bp, compared with standards [30 bp oligo (IDT)/100 bp DNA ladder (NEB)], were excised, frozen, and then purified using a Freeze ‘N Squeeze gel purification kit (BioRad) because the small band size precluded spin column purification. Purified digested fragments were mixed together at a 1:1 ratio and assembled via overlap-extension PCR.
Generating a Multiple Sequence Alignment and Computing a Consensus Sequence.
We searched various resources including GenBank, SwisProt, UniProt, NCBI-BLAST, and patent databases for reported FP sequences. We found 741 unique fluorescent protein sequences and aligned them with MAFFT, which we then hand-curated with the use of a 163-member structural alignment. Each position in the resulting alignment was scored as follows (31, 52). First, sequences were Henikoff weighted (53) to account for the presence of highly similar sequences in the MSA. The entropy of each column in the alignment was then calculated, ignoring any gaps. Finally, a score was obtained for each nongap character in each column via
Here is the frequency of character in column , and is the uncertainty reduction in each column [corrected for the column gap fraction ] calculated as
The highest scoring character in each column (or multiple characters in the case of ties) was selected as the consensus amino acid at that position.
Protein Expression and Library Screening.
Single bacterial colonies were picked with sterile toothpicks and inoculated into 300 µL of Super Optimal Broth (SOB) supplemented with 100 µg/mL ampicillin in 2 mL deep-well 96-well plates (Seahorse Biosciences). The plates were sealed with microporous film (Denville Scientific) to facilitate gas exchange during growth. Cultures were grown overnight at 37 °C/300 rpm in an Infors-HT Multitron shaking incubator. The next morning, 800 µL of fresh SOB with 100 µg/mL ampicillin and 1 mM Isopropyl β-d-1-thiogalactopyranoside (IPTG) was added to a total volume of 1 mL (evaporation losses overnight are ∼100 µL). Plates were then shaken 12 h at either 30 °C or 37 °C and 400 rpm. After overnight expression, plates were screened with a liquid handling robot (Tecan Freedom Evo) linked to a platereader (Tecan Saffire 2). Two hundred µL of each culture was added to Greiner UV-Star 96-well plates and imaged for fluorescence emission at 675 nm after excitation at 600 nm. Controls were included on each plate to account for plate-to-plate variation. Potential hits were streaked out onto a fresh LB-Amp plate, grown overnight at 37 °C, and four colonies were picked for each potential hit. These were then grown again and screened as detailed above, with hits then ranked on their significant variation from the parent or control.
Protein Purification.
To further characterize important variants, 1 L of SOB in Fernbach flasks was inoculated 1:100 with overnight cultures, grown to an OD of ∼0.5, and induced at 37 °C for 12 h with 1 mM IPTG. The broth was then transferred to centrifuge flasks and spun at 5,000 × g in a fixed angle rotor for 10 min and the supernatant decanted. Bacterial pellets were resuspended in 25 mL of lysis buffer (50 mM sodium phosphate, 150 mM NaCl, 0.1% vol/vol Triton-X, pH 7.4) supplemented with 50 units/mL Benzonase (Sigma) and 0.05 mg/mL Hen Egg Lysozyme (Sigma). Resuspended pellets were then run over a microfluidizer to fully lyse the bacteria. To pellet down the cellular debris, the lysed cultures were again centrifuged for 10 min at 15,000 × g in a fixed angle rotor. The colored supernatant was then poured through a column of His-Select resin (Sigma), washed twice (50 mM sodium phosphate, 150 mM NaCl, 15 mM Imidazole, pH 7.4), and eluted with 500 µL elution buffer (50 mM sodium phosphate, 150 mM NaCl, 250 mM Imidazole, pH 7.4). Proteins were further purified by SEC (AKTA) with a Superdex 75 10/300 column and in the process buffer exchanged into PBS.
Fluorescent Protein Characterization.
Purified protein variants were assayed in triplicate in Greiner UV-Star 96-well plates with a Tecan Saffire 2. An absorbance scan (260–650 nm), a fluorescence excitation scan (500–640 nm excitation/675 nm emission), and a fluorescence emission scan (550 nm excitation/575–800 nm emission) were run on 100 µL of eluted protein to determine spectral peaks.
To measure the quantum yield we diluted each protein so that the absorbance for 200 µL of protein at 540 nm was between 0.1 and 0.5. We then measured the A550 in triplicate (or duplicate if it was a poorly expressed protein), diluted the sample to an A550 of 0.04, and took an emission scan (540 nm excitation/550–800 nm emission). The area under the emission curve was calculated after fitting it to a fourth-order Gaussian, and the quantum yield was calculated with the following formula:
where Φ is quantum yield, A is absorbance, F is total fluorescent emission (area under the curve), and n is the refractive index of the solvents used. Subscript X refers to the queried substance, and subscript S refers to a standard of known quantum yield. It is important that the standard be excited with the same wavelength of light as the unknown sample. We use DsRed, which has a known quantum yield of 0.79 as the protein standard.
To measure extinction coefficient we took 100 µl of the protein solution that had been diluted to an A550 of between 0.1 and 0.5 and measured absorbance between 400 nm and 700 nm in triplicate. We then added 100 µl of 2 M NaOH to each well and remeasured absorbance between 400 nm and 700 nm. The base-denatured chromophore, which peaks at ∼450 nm has a known extinction coefficient of 44,000 M−1cm−1. Then to calculate the extinction coefficient is calculated with the following formula:
Photobleaching kinetics were measured by imaging aqueous droplets of purified protein in mineral oil using an Axiovert 200M (Zeiss) equipped with a 75-W xenon-arc lamp, a 40× objective lens (NA = 1.3, oil), and a digital CMOS camera (Orca flash 4.0; Hamamatsu) and controlled by Metamorph software (Molecular Devices). Images of protein droplets (n = 3) were taken every 30 s under continuous illumination with a 565/50 nm excitation filter (Semrock) for HcRed/mGinger1/mGinger2 and a 605/55 nm excitation filter (Semrock) for mCardinal/mKelly1/mKelly2.
Chromophore maturation was measured in a BL21(DE3) Escherichia coli strain. Transformed bacteria were grown at 37 °C in sealed N2-filled culture tubes overnight for anaerobic protein expression. Bacteria were then harvested by centrifugation and lysed by B-PER (Thermo). Lysate (n = 3) fluorescence were monitored as a function of time on the Safire2 plate reader (Tecan) at 37 °C.
To characterize pH sensitivity, fluorescence intensity as a function of pH was determined by dispensing purified protein in buffer (n = 3) into a 384-well plate. Measurements were taken in a Safire2 plate reader (Tecan). pH buffer solutions from pH 3 to 11 were prepared according to the Carmody buffer system (54).
Thermal Stability.
Purified proteins were diluted to an absorbance of 0.2 at the wavelength of maximum absorbance (λabs) so that their fluorescence would not saturate the rtPCR detector. Fifty µL of each purified protein was then loaded into a 96-well PCR plate and covered with clear optical tape. The proteins were incubated at 37 °C for 10 min, and then the temperature was ramped at 0.5 °C every 30 s up to 99 °C, with fluorescence measured every ramp step in a CFX96 Touch Real-Time PCR Detection System (Bio-Rad). We refer to this as a thermal melt. The derivative curve of the thermal melt finds the inflection point of the slope, which is the apparent temperature at which fluorescence is irrevocably lost (apparent Tm).
Oligomeric Determination.
Size exclusion chromatography.
One hundred µL of each purified protein analyzed was run over a Superdex 75 10/300 size exclusion column with 25 mL bed volume on an AKTA from GE Life Sciences. Absorbance was measured after passage through the column at 575 nm, where the red chromophore absorbs.
Analytical ultracentrifugation.
Purified protein samples were diluted to an A575 of 0.5 for a path length of 1.25 cm. These samples were put into two-channel sedimentation velocity cuvettes with the blank channel containing PBS. Sedimentation velocity was run at 40,000 rpm overnight with full A575 scans collected with no pause between reads. Data were loaded into Sedfit, and a c(m) distribution was run with default assumptions made for PBS buffer viscosity. After integration, the c(m) curve was exported to Excel.
CytERM constructs.
CytERM constructs were constructed for HcRed, mGinger1, mGinger2, mKelly1, and mKelly2 amplified with a 5′ primer with an AgeI site and a 3′ primer with a NotI site. The purified PCR products were then digested and ligated into a similarly digested CytERM-mGFP (Plasmid 62237; Addgene). HeLa cells were maintained in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10% FBS (Sigma) and Glutamax (Life Technologies) and incubated at 37 °C with 5% CO2. Transient transfections were performed using Turbofect (Thermo Scientific) according to the manufacturer’s guidelines. Transfected cells were imaged using an Axiovert 200M (Zeiss) equipped with a 75-W xenon-arc lamp, a 40× objective lens (NA = 1.3, oil), and a digital CMOS camera (Orca flash 4.0; Hamamatsu) and driven by Metamorph software (Molecular Devices).
Crystallography.
Rectangular plate crystals of HcRed7 grew in 7 d by the sitting-drop vapor diffusion method in 100 mM Bis-Tris, pH 6.5, with 200 mM ammonium sulfate and 25% wt/vol PEG 3350. Crystals were flash frozen in 2-Methyl-2,4-pentanediol (MPD) and shipped to beamline 12-2 at the Stanford Synchrotron Radiation Lightsource, where a 1.63-Å dataset was collected. Phases were obtained through molecular replacement using the crystal structure of HcRed (PDB ID 1YZW). Following molecular replacement, model building and refinement were run with COOT and PHENIX (55, 56). NCS restraints were applied to early refinement steps and removed at the final stages of refinement. TLS parameters were used throughout. The chromophore was initially left out of the refinement and added at a later stage when clear density became evident for it. Coordinates were deposited in the Protein Data Bank with the code 6DEJ. Data collection and refinement statistics are listed in SI Appendix, Table S6.
Supplementary Material
Acknowledgments
We thank Yun Mou, Matthew Moore, and Roberto Chica for many helpful conversations. We would also like to thank Prof. Frances Arnold for her advice and support. Jens Kaiser and Pavle Nikolovski were generous with their time and advice in troubleshooting protein crystallography. The authors are grateful for the use of the beamline 12-2 at the Stanford Synchrotron Radiation Lightsource and to the Gordon and Betty Moore Foundation for support of the Molecular Observatory at the California Institute of Technology. We also acknowledge the funding and support of the National Institute of Biomedical Imaging and Bioengineering (Grant R21EB018579).
Footnotes
The authors declare no conflict of interest.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org (PDB ID code: 6DEJ). GenBank IDs [accession nos. MK040729 (mGinger), MK040730 (mGinger2), MK040731 (mKelly1), and MK040732 (mKelly2)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807449115/-/DCSupplemental.
References
- 1.Tromberg BJ, et al. Non-invasive in vivo characterization of breast tumors using photon migration spectroscopy. Neoplasia. 2000;2:26–40. doi: 10.1038/sj.neo.7900082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bashkatov AN, Genina EA, Kochubey VI, Tuchin VV. Optical properties of human skin, subcutaneous and mucous tissues in the wavelength range from 400 to 2000 nm. J Phys D Appl Phys. 2005;38:2543–2555. [Google Scholar]
- 3.Wang S, Moffitt JR, Dempsey GT, Xie XS, Zhuang X. Characterization and development of photoactivatable fluorescent proteins for single-molecule–based superresolution imaging. Proc Natl Acad Sci USA. 2014;111:8452–8457. doi: 10.1073/pnas.1406593111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang W, Li GW, Chen C, Xie XS, Zhuang X. Chromosome organization by a nucleoid-associated protein in live bacteria. Science. 2011;333:1445–1449. doi: 10.1126/science.1204697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Filonov GS, et al. Bright and stable near-infrared fluorescent protein for in vivo imaging. Nat Biotechnol. 2011;29:757–761. doi: 10.1038/nbt.1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shcherbakova DM, et al. Bright monomeric near-infrared fluorescent proteins as tags and biosensors for multiscale imaging. Nat Commun. 2016;7:12405. doi: 10.1038/ncomms12405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Strack RL, Strongin DE, Mets L, Glick BS, Keenan RJ. Chromophore formation in DsRed occurs by a branched pathway. J Am Chem Soc. 2010;132:8496–8505. doi: 10.1021/ja1030084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Piatkevich KD, et al. Extended Stokes shift in fluorescent proteins: Chromophore-protein interactions in a near-infrared TagRFP675 variant. Sci Rep. 2013;3:1847. doi: 10.1038/srep01847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Z, et al. Mutagenesis of mNeptune red-shifts emission spectrum to 681-685 nm. PLoS One. 2016;11:e0148749. doi: 10.1371/journal.pone.0148749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Alieva NO, et al. Diversity and evolution of coral fluorescent proteins. PLoS One. 2008;3:e2680. doi: 10.1371/journal.pone.0002680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shagin DA, et al. GFP-like proteins as ubiquitous metazoan superfamily: Evolution of functional features and structural complexity. Mol Biol Evol. 2004;21:841–850. doi: 10.1093/molbev/msh079. [DOI] [PubMed] [Google Scholar]
- 12.Bou-Abdallah F, Chasteen ND, Lesser MP. Quenching of superoxide radicals by green fluorescent protein. Biochim Biophys Acta. 2006;1760:1690–1695. doi: 10.1016/j.bbagen.2006.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Palmer CV, Modi CK, Mydlarz LD. Coral fluorescent proteins as antioxidants. PLoS One. 2009;4:e7298. doi: 10.1371/journal.pone.0007298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salih A, Larkum A, Cox G, Kühl M, Hoegh-Guldberg O. Fluorescent pigments in corals are photoprotective. Nature. 2000;408:850–853. doi: 10.1038/35048564. [DOI] [PubMed] [Google Scholar]
- 15.Smith EG, D’Angelo C, Salih A, Wiedenmann J. Screening by coral green fluorescent protein (GFP)-like chromoproteins supports a role in photoprotection of zooxanthellae. Coral Reefs. 2013;32:463–474. [Google Scholar]
- 16.Campbell RE, et al. A monomeric red fluorescent protein. Proc Natl Acad Sci USA. 2002;99:7877–7882. doi: 10.1073/pnas.082243699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kredel S, et al. mRuby, a bright monomeric red fluorescent protein for labeling of subcellular structures. PLoS One. 2009;4:e4391. doi: 10.1371/journal.pone.0004391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shemiakina II, et al. A monomeric red fluorescent protein with low cytotoxicity. Nat Commun. 2012;3:1204. doi: 10.1038/ncomms2208. [DOI] [PubMed] [Google Scholar]
- 19.Kogure T, et al. A fluorescent variant of a protein from the stony coral Montipora facilitates dual-color single-laser fluorescence cross-correlation spectroscopy. Nat Biotechnol. 2006;24:577–581. doi: 10.1038/nbt1207. [DOI] [PubMed] [Google Scholar]
- 20.Bindels DS, et al. mScarlet: A bright monomeric red fluorescent protein for cellular imaging. Nat Methods. 2017;14:53–56. doi: 10.1038/nmeth.4074. [DOI] [PubMed] [Google Scholar]
- 21.Gurskaya NG, et al. GFP-like chromoproteins as a source of far-red fluorescent proteins. FEBS Lett. 2001;507:16–20. doi: 10.1016/s0014-5793(01)02930-1. [DOI] [PubMed] [Google Scholar]
- 22.Subach OM, et al. Conversion of red fluorescent protein into a bright blue probe. Chem Biol. 2008;15:1116–1124. doi: 10.1016/j.chembiol.2008.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wannier TM, Moore MM, Mou Y, Mayo SL. Computational design of the β-sheet surface of a red fluorescent protein allows control of protein oligomerization. PLoS One. 2015;10:e0130582. doi: 10.1371/journal.pone.0130582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wall MA, Socolich M, Ranganathan R. The structural basis for red fluorescence in the tetrameric GFP homolog DsRed. Nat Struct Biol. 2000;7:1133–1138. doi: 10.1038/81992. [DOI] [PubMed] [Google Scholar]
- 25.Wannier TM, Mayo SL. The structure of a far-red fluorescent protein, AQ143, shows evidence in support of reported red-shifting chromophore interactions. Protein Sci. 2014;23:1148–1153. doi: 10.1002/pro.2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mudalige K, et al. Photophysics of the red chromophore of HcRed: Evidence for cis-trans isomerization and protonation-state changes. J Phys Chem B. 2010;114:4678–4685. doi: 10.1021/jp9102146. [DOI] [PubMed] [Google Scholar]
- 27.Wilmann PG, et al. The 2.1 Å crystal structure of the far-red fluorescent protein HcRed: Inherent conformational flexibility of the chromophore. J Mol Biol. 2005;349:223–237. doi: 10.1016/j.jmb.2005.03.020. [DOI] [PubMed] [Google Scholar]
- 28.Moore MM, Oteng-Pabi SK, Pandelieva AT, Mayo SL, Chica RA. Recovery of red fluorescent protein chromophore maturation deficiency through rational design. PLoS One. 2012;7:e52463. doi: 10.1371/journal.pone.0052463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bloom JD, Labthavikul ST, Otey CR, Arnold FH. Protein stability promotes evolvability. Proc Natl Acad Sci USA. 2006;103:5869–5874. doi: 10.1073/pnas.0510098103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jäckel C, Bloom JD, Kast P, Arnold FH, Hilvert D. Consensus protein design without phylogenetic bias. J Mol Biol. 2010;399:541–546. doi: 10.1016/j.jmb.2010.04.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schneider TD. Consensus sequence Zen. Appl Bioinformatics. 2002;1:111–119. [PMC free article] [PubMed] [Google Scholar]
- 32.Pletneva NV, et al. Crystallographic study of red fluorescent protein eqFP578 and its far-red variant Katushka reveals opposite pH-induced isomerization of chromophore. Protein Sci. 2011;20:1265–1274. doi: 10.1002/pro.654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Piatkevich KD, Malashkevich VN, Almo SC, Verkhusha VV. Engineering ESPT pathways based on structural analysis of LSSmKate red fluorescent proteins with large Stokes shift. J Am Chem Soc. 2010;132:10762–10770. doi: 10.1021/ja101974k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Costantini LM, Fossati M, Francolini M, Snapp EL. Assessing the tendency of fluorescent proteins to oligomerize under physiologic conditions. Traffic. 2012;13:643–649. doi: 10.1111/j.1600-0854.2012.01336.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Crameri A, Whitehorn EA, Tate E, Stemmer WP. Improved green fluorescent protein by molecular evolution using DNA shuffling. Nat Biotechnol. 1996;14:315–319. doi: 10.1038/nbt0396-315. [DOI] [PubMed] [Google Scholar]
- 36.Zhao H, Arnold FH. Optimization of DNA shuffling for high fidelity recombination. Nucleic Acids Res. 1997;25:1307–1308. doi: 10.1093/nar/25.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Regmi CK, Bhandari YR, Gerstman BS, Chapagain PP. Exploring the diffusion of molecular oxygen in the red fluorescent protein mCherry using explicit oxygen molecular dynamics simulations. J Phys Chem B. 2013;117:2247–2253. doi: 10.1021/jp308366y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Black SD, Mould DR. Development of hydrophobicity parameters to analyze proteins which bear post- or cotranslational modifications. Anal Biochem. 1991;193:72–82. doi: 10.1016/0003-2697(91)90045-u. [DOI] [PubMed] [Google Scholar]
- 39.Chennamsetty N, Voynov V, Kayser V, Helk B, Trout BL. Prediction of aggregation prone regions of therapeutic proteins. J Phys Chem B. 2010;114:6614–6624. doi: 10.1021/jp911706q. [DOI] [PubMed] [Google Scholar]
- 40.Goedhart J, et al. Structure-guided evolution of cyan fluorescent proteins towards a quantum yield of 93% Nat Commun. 2012;3:751. doi: 10.1038/ncomms1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lelimousin M, et al. Intrinsic dynamics in ECFP and cerulean control fluorescence quantum yield. Biochemistry. 2009;48:10038–10046. doi: 10.1021/bi901093w. [DOI] [PubMed] [Google Scholar]
- 42.Rurack K, Spieles M. Fluorescence quantum yields of a series of red and near-infrared dyes emitting at 600-1000 nm. Anal Chem. 2011;83:1232–1242. doi: 10.1021/ac101329h. [DOI] [PubMed] [Google Scholar]
- 43.Ren H, et al. Cysteine sulfoxidation increases the photostability of red fluorescent proteins. ACS Chem Biol. 2016;11:2679–2684. doi: 10.1021/acschembio.6b00579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chica RA, Moore MM, Allen BD, Mayo SL. Generation of longer emission wavelength red fluorescent proteins using computationally designed libraries. Proc Natl Acad Sci USA. 2010;107:20257–20262. doi: 10.1073/pnas.1013910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Morozova KS, et al. Far-red fluorescent protein excitable with red lasers for flow cytometry and superresolution STED nanoscopy. Biophys J. 2010;99:L13–L15. doi: 10.1016/j.bpj.2010.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Grigorenko BL, Nemukhin AV, Polyakov IV, Krylov AI. Triple-decker motif for red-shifted fluorescent protein mutants. J Phys Chem Lett. 2013;4:1743–1747. doi: 10.1021/jz4006288. [DOI] [PubMed] [Google Scholar]
- 47.Tubbs JL, Tainer JA, Getzoff ED. Crystallographic structures of Discosoma red fluorescent protein with immature and mature chromophores: Linking peptide bond trans-cis isomerization and acylimine formation in chromophore maturation. Biochemistry. 2005;44:9833–9840. doi: 10.1021/bi0472907. [DOI] [PubMed] [Google Scholar]
- 48.Pandelieva AT, et al. Brighter red fluorescent proteins by rational design of triple-decker motif. ACS Chem Biol. 2015;11:508–517. doi: 10.1021/acschembio.5b00774. [DOI] [PubMed] [Google Scholar]
- 49.Matela G, et al. A far-red emitting fluorescent marker protein, mGarnet2, for microscopy and STED nanoscopy. Chem Commun (Camb) 2017;53:979–982. doi: 10.1039/c6cc09081h. [DOI] [PubMed] [Google Scholar]
- 50.Strongin DE, et al. Structural rearrangements near the chromophore influence the maturation speed and brightness of DsRed variants. Protein Eng Des Sel. 2007;20:525–534. doi: 10.1093/protein/gzm046. [DOI] [PubMed] [Google Scholar]
- 51.Henderson JN, et al. Excited state proton transfer in the red fluorescent protein mKeima. J Am Chem Soc. 2009;131:13212–13213. doi: 10.1021/ja904665x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schneider TD, Stephens RM. Sequence logos: A new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henikoff S, Henikoff JG. Position-based sequence weights. J Mol Biol. 1994;243:574–578. doi: 10.1016/0022-2836(94)90032-9. [DOI] [PubMed] [Google Scholar]
- 54.Carmody WR. Easily prepared wide range buffer series. J Chem Educ. 1961;38:559. [Google Scholar]
- 55.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.