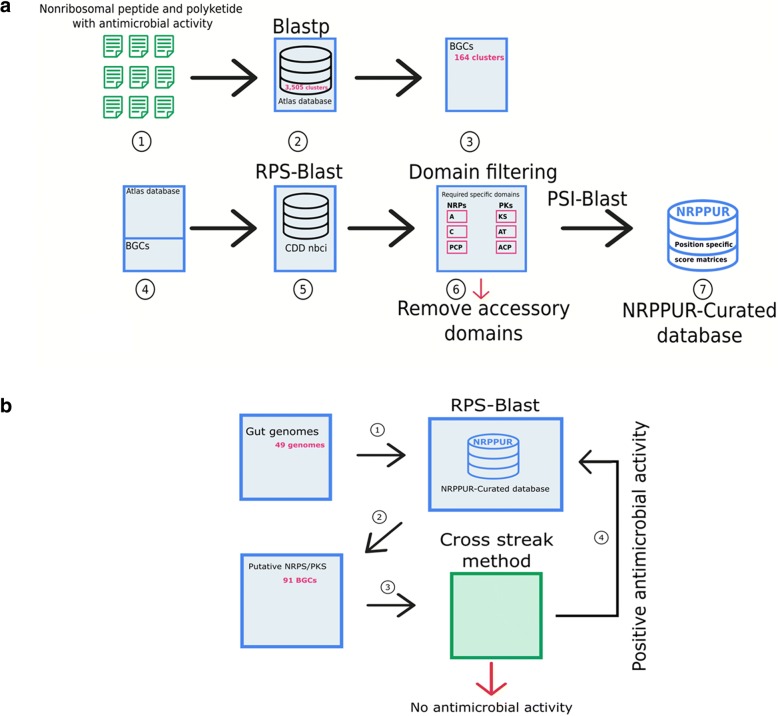
Fig. 1.
NRPPUR database construction and utilization. a Flowchart depicting the elaboration of NRPPUR DB. 1- Literature scan to find NRPS/PKS clusters with experimentally evidence of antimicrobial activity, 2- BLASTp analysis against Atlas database and removal of duplicate sequences, 3- Recuperation of 164 Biosynthetic Gene Clusters, 4- Assembly of the 164 BGCs to the existing Atlas database, 5- Identification of NRPS/PKS domains using RPS-BLAST against Conserved Domain Database, 6- Conservation of the minimal functional domains adenylation (A), condensation (C) and peptidyl carrier protein (PCP) in the case of NRPS and acyltransferase (A) domain, an acyl carrier protein (ACP) and a ketoacyl synthase (KS) domains in the case of PKS, 7- Elaboration of Position Specific Score Matrices for each domain using PSI-BLAST. b Schematic diagram representing the different steps for detection of NRPS/PKS clusters. 1- Analysis of 49 gut genomes using RPS-BLAST against NRPPUR DB, 2- Identification of putative NRPS/PKS clusters, 3- In vitro verification of antimicrobial activity using Cross-streak method, 4- Database increment with BGCs from species showing positive antimicrobial activity. The in silico steps are shown in blue and the experimental in vitro steps in green