Abstract
The identity of the gonads is determined by which fate, ovarian granulosa cell or testicular Sertoli cell, the bipotential somatic cell precursors choose to follow. In most vertebrates, the conserved transcription factor FOXL2 contributes to the fate of granulosa cells. To understand FOXL2 functions during gonad differentiation, we performed genome-wide analysis of FOXL2 chromatin occupancy in fetal ovaries and established a genetic mouse model that forces Foxl2 expression in the fetal testis. When FOXL2 was ectopically expressed in the somatic cell precursors in the fetal testis, FOXL2 was sufficient to repress Sertoli cell differentiation, ultimately resulting in partial testis-to-ovary sex-reversal. Combining genome-wide analysis of FOXL2 binding in the fetal ovary with transcriptomic analyses of our Foxl2 gain-of-function and previously published Foxl2 loss-of-function models, we identified potential pathways responsible for the feminizing action of FOXL2. Finally, comparison of FOXL2 genome-wide occupancy in the fetal ovary with testis-determining factor SOX9 genome-wide occupancy in the fetal testis revealed extensive overlaps, implying that antagonistic signals between FOXL2 and SOX9 occur at the chromatin level.
Introduction
The signals that trigger sex determination of the gonads vary widely among vertebrate species, from environmental determinants in some turtles and alligators to chromosomal composition in birds and humans. Despite these differences, a relatively conserved set of factors operates to induce and/or maintain gonadal differentiation, such as DMRT1 and SOX genes in the testis and FOXL2, CYP19A1 and WNT4/RSPO1 in the ovary (1). A link between the transcription factor FOXL2 and ovarian development in humans was initially identified in women suffering from type I Blepharophymosis, Ptosis and Epicanthus inversus Syndrome. This syndrome, caused by a heterozygous dominant mutation in the FOXL2 gene, results in primary ovarian insufficiency (2). Ovarian expression of FOXL2 has now been reported in most classes of vertebrate species, and even in invertebrates (3). From fish to human, FOXL2 is one of the most conserved regulators of ovarian granulosa cell identity. Loss of FOXL2 results in embryonic ovary-to-testis sex reversal in fish and goat (4,5). In mice, FOXL2 is expressed in ovarian granulosa cells from sex determination to adulthood (6). However, contrary to fish and goat, global loss of Foxl2 in mice only causes sex-reversal postnatally (7–9). Meanwhile, conditional deletion of Foxl2 in adult mouse ovary leads to transdifferentiation of granulosa cells into Sertoli cells, indicating that FOXL2 is required for fate maintenance of granulosa cells (10). These observations lead to the hypotheses that in the mouse fetal ovary, FOXL2 has limited roles in the control of granulosa cell differentiation, or redundant action of other factors in addition to FOXL2 contributes to granulosa cell differentiation. Multiple mouse genetic models support the second hypothesis. Indeed, combined loss of Foxl2/Wnt4 or Foxl2/Rspo1 genes in XX embryos produces a more pronounced ovary-to-testis sex reversal phenotype than each single knockout (11,12), demonstrating a complementary role of FOXL2 and the Wingless-type mouse mammary tumor virus integration site (Wnt)/beta-catenin pathway in granulosa cell differentiation. A role of FOXL2 in mouse gonadal differentiation was further supported by the anti-testis properties of FOXL2 in male transgenic mouse embryos with ubiquitous FOXL2 expression (12,13).
In this study, we aimed to understand how FOXL2 alters the transcriptional landscape in the fetal gonads and tips the balance toward granulosa cell fate. We combined three approaches: first, we identified the genome-wide FOXL2-bound chromatin regions in the fetal ovary by ChIP-seq and determined the gene regulatory networks potentially controlled by FOXL2. Second, we generated a Foxl2 gain-of-function model to investigate whether ectopic presence of FOXL2 in the fetal testis is sufficient to drive supporting cell differentiation into granulosa cells. Third, by comparing the potential direct target genes of FOXL2 identified by ChIP-seq in the fetal ovary with the transcriptomic changes from published Foxl2 loss-of-function and our Foxl2 gain-of-function models, we uncovered biologically relevant potential targets downstream of FOXL2 that contribute to the control of supporting cell fate in the gonads.
Results
Genome-wide identification of FOXL2 chromatin binding sites in the fetal ovary
To better understand the molecular action of FOXL2 in controlling supporting cell identity, we performed genome-wide chromatin immunoprecipitation followed by sequencing (ChIP-seq) on chromatin from pools of fetal ovaries collected at embryonic day E14.5 (Fig. 1; Supplementary Material, Dataset S1). The specificity of the FOXL2 antibody was confirmed by immunofluorescences on control newborn gonads as well as on gonads from both Foxl2 loss-of-function and gain-of-function mouse models (Supplementary Material, Fig. S1A). FOXL2 ChIP-seq in the fetal ovary led to the identification of 11 438 peaks. The best match for the top de novo motif identified by HOMER (6.2-fold enrichment compared to the background, P-value 1e-685) corresponded to the consensus FOXL2 DNA motif previously identified in the adult granulosa cells (GSE60858; 14; Fig. 1A). The majority of the peaks were located either within the gene body (26% exon; 24% intron; Fig. 1B) or close to the TSS (25% Promoter: <1 kb of TSS; 12% Upstream: −10 to −1 kb of TSS; Fig. 1B). FOXL2 peaks were assigned to the nearest gene, resulting in 9324 potential direct target genes of FOXL2 (purple circle in Fig. 1C). We then used EnrichR, a comprehensive gene set enrichment analysis tool (15,16), for pathway analyses and found enrichment in FOXO signaling, a pathway linked to cell-cycle arrest and longevity (17) and multiple other pathways related to cell cycle, cancer or stem cell pluripotency (Supplementary Materials, Fig. S1B and Dataset S1).
Figure 1.
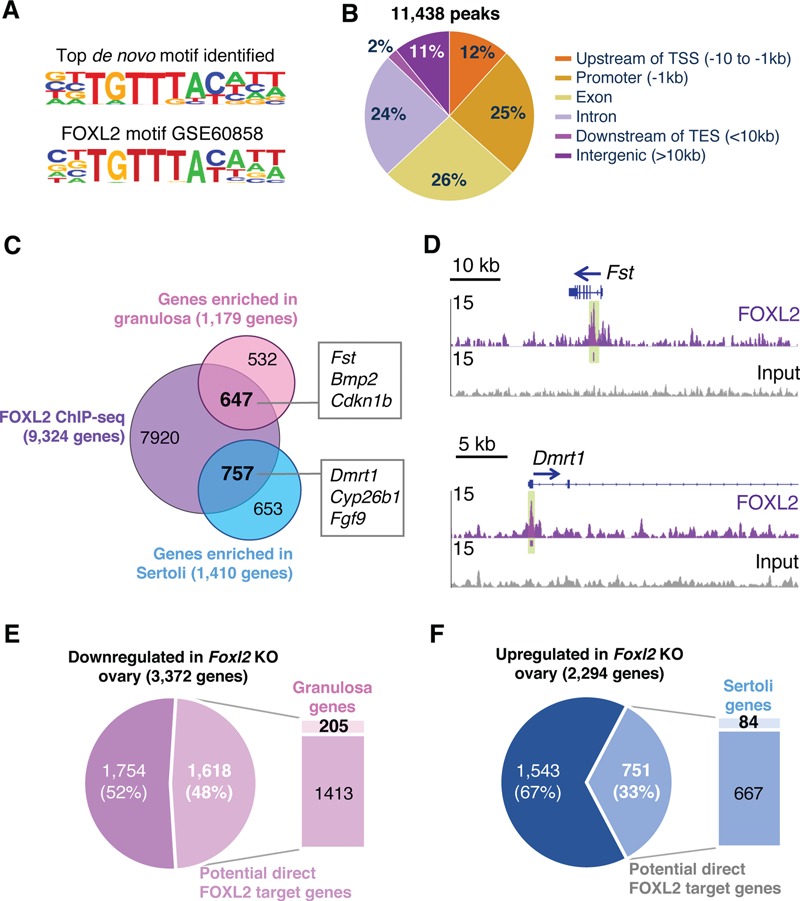
Genome-wide identification of FOXL2-binding regions in the ovary. (A) The top motif identified by HOMER de novo analysis (P-value 1e-685) for FOXL2 ChIP-seq in E14.5 ovaries corresponds to the published FOXL2 binding sequence (14). (B) Distribution of genomic locations of the 11 438 FOXL2 binding peaks. TSS: Transcription Start Site; TES: Transcription End Site. (C) Venn diagram comparing FOXL2 potential direct target genes in the fetal ovary (ChIP-seq) with genes differentially expressed in E13.5 granulosa versus Sertoli cells (18). (D) Genome browser view of a gene enriched in granulosa cells (Fst) and a gene enriched in Sertoli cells (Dmrt1) bound by FOXL2 in the fetal ovary. Arrows: gene orientation; Green highlights: significant FOXL2-binding peaks identified by HOMER. (E and F) Identification of the genes downregulated (E) or upregulated (F) in Foxl2 knockout (KO) ovaries (13) that are bound by FOXL2 and enriched in fetal granulosa or Sertoli cells, respectively.
Identification of potential target genes of FOXL2 in the fetal supporting cell lineage
FOXL2 is expressed in the supporting cell lineage in the ovary but not in their testicular counterparts (6); therefore, we could assume that potential direct target genes of FOXL2 are expressed in a sexually dimorphic manner between female and male supporting cells. In order to focus on the supporting cell lineage, we leveraged the published transcriptomic data from isolated cell populations of the mouse fetal gonads (18), and used them to identify genes expressed differentially between fetal granulosa cells and their male counterpart Sertoli cells (Fig. 1C; Supplementary Material, Dataset S2). Comparison of FOXL2 ChIP-seq data with the genes enriched in fetal granulosa cells (1179 genes in pink circle in Fig. 2C) or Sertoli cells (1410 genes in blue circle in Fig. 2C) allowed us to identify the potential FOXL2 target genes that could contribute to supporting cell differentiation. Respectively, 55% (647) of the genes enriched in granulosa cells and 54% (757) of the genes enriched in Sertoli cells were potential direct target of FOXL2. Among them were the granulosa genes Fst and Bmp2 and the Sertoli genes Dmrt1, Cyp26b1 and Fgf9 (Fig. 1C). For instance, FOXL2 binding peaks were located in the first intron of Fst gene as previously identified (14,19) and at the promotor of Dmrt1 gene (Fig. 1D).
Figure 2.
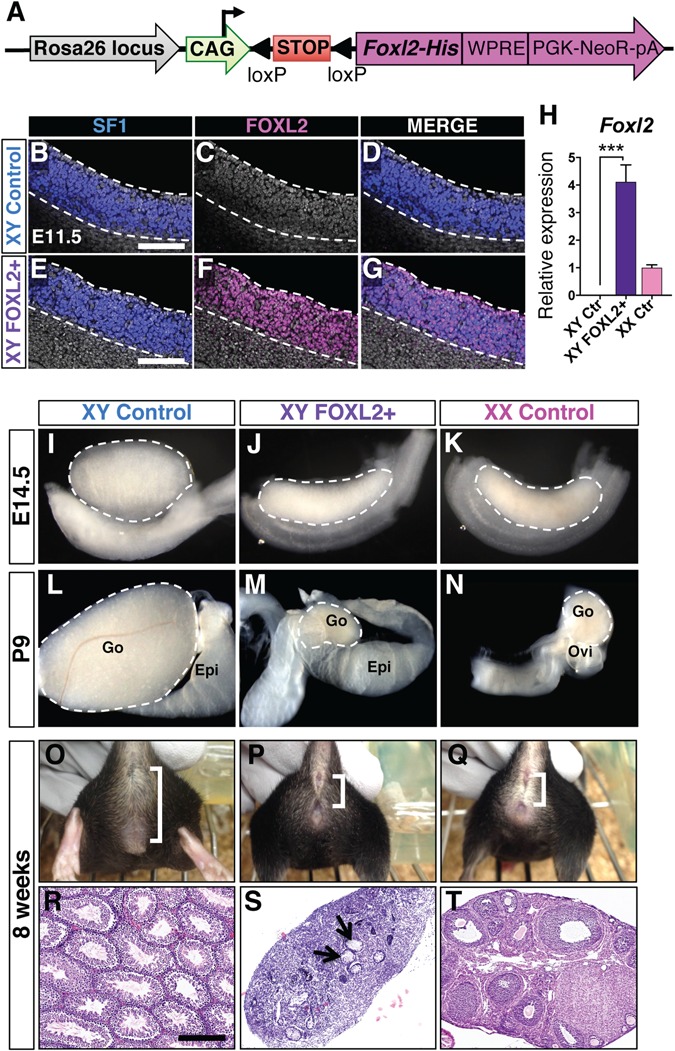
Ectopic induction of FOXL2 expression in XY fetal gonads results in partial male-to-female sex reversal. (A) Design of Rosa26-CAG-LSL-Foxl2 construct. Cre-mediated recombination excises the LoxP-flanked STOP cassette (LSL), resulting in expression of FOXL2 in Cre-expressing cells. His: His-Tag. WPRE: Woodchuck Hepatitis Virus Posttranscriptional Regulatory Element. pA: polyadenylation signal. (B–G) Whole mount immunofluorescences for SF1 and FOXL2 in XY control and FOXL2+ E11.5 gonads show ectopic FOXL2 expression in the SF1+ cells. Nuclei were stained with DAPI (gray). Dotted lines outline the gonads. Scale bars: 150 μm. (H) qPCR analyses of Foxl2 expression in E14.5 gonads. t-test; mean ± SEM (n = 4); ***P < 0.001. Controls: littermates of XY FOXL2+ embryos lacking the Sf1-Cre transgene. (I–N) Bright field images of XY control, XY FOXL2+ and XX control gonads and reproductive tracts at E14.5 (I–K) and P9 (L–N). Epi: epididymis; Go: gonad; Ovi: oviduct. Dashed lines outline gonads. (O–Q) External genitalia of 8-week-old mice. Brackets: anogenital distance. (R–T) H&E stained sections of 8-week-old gonads. Arrowheads: rudimentary seminiferous tubules. Scale bar: 300 μm.
To establish the functional relevance of FOXL2 chromatin binding in the fetal ovary, we next compared the potential FOXL2 direct target genes identified in our ChIP-seq with the transcriptome of ovaries lacking Foxl2. We hypothesized that expression of FOXL2 direct target genes would be affected significantly in the absence of Foxl2. The phenotype of Foxl2 KO ovaries has been extensively studied by others (7–9,12,13): Although loss of Foxl2 led to sex reversal only after birth, significant changes in ovarian transcriptome were detected before birth. We obtained the publicly available lists of genes expressed differentially between the control and Foxl2 KO fetal/neonatal ovaries (13), and compared them with our ChIP-seq gene list (Fig. 1E–F and Supplementary Material, Dataset S2). We found that among the 3372 genes that were downregulated in the Foxl2 KO ovary (Fig. 1E), 48% (1618 genes) were identified in our FOXL2 ChIP-seq experiment. Among them, 205 genes were enriched in fetal granulosa cells, representing candidates for positive regulation by FOXL2 in the fetal ovary. For the 2294 genes that were upregulated in the Foxl2 KO ovary (Fig. 1F), 33% (751 genes) were identified in our FOXL2 ChIP-seq experiment (Fig. 1F). Among them, 84 genes were enriched in the fetal Sertoli cells, representing candidates that are repressed by FOXL2 in the fetal ovary. Wt1 gene was one of the genes upregulated in the absence of Foxl2 (Supplementary Material, Fig. S1C): in addition to a FOXL2 peak found near the Wt1 transcription starting site, another FOXL2-bound region was found in the newly identified gonad-specific Wt1 enhancer region (20). A transgenic reporter model for this enhancer demonstrated that this regulatory region drives Wt1 expression in the Sertoli cells of the fetal testis. These results suggest that FOXL2 could contribute to regulation of Wt1 expression in the gonadal supporting cells.
Ectopic FOXL2 expression in the fetal testis results in partial male-to-female sex reversal
The upregulation of genes normally expressed in Sertoli cells in the Foxl2 KO ovary implicates an anti-testis property of FOXL2 (Fig. 1F). In addition, XY mice with ubiquitous FOXL2 expression in response to heat shock developed malformed testes and aberrant gene expression at E13.5 (12,13). However, this model was limited by the ubiquitous nature of FOXL2 induction, the complication of heat shock, the unknown timing of ectopic FOXL2 expression and the absence of whole-genome transcriptomic analyses of the gonads. In order to identify the specific impact of FOXL2 in the somatic cells of the fetal testis, we generated a new model that enabled us to control the cell type and timing of ectopic FOXL2 expression. We integrated the targeting construct CAG-LSL-Foxl2 into the Rosa26 locus by homologous recombination (Fig. 2A and Supplementary Material, Fig. S2). In the presence of Cre recombinase, the loxP-flanked STOP cassette was removed, therefore allowing FOXL2 expression in the Cre-expressing cells. The Rosa26-CAG-LSL-Foxl2 mice were crossed to the Sf1-Cre mice, a validated Cre model for targeting recombination in the somatic cells of fetal gonads around E10.5–11.5, the onset of sex determination (21–25). We confirmed that FOXL2 expression was induced in the Sf1-Cre;Rosa26-CAG-LSL-Foxl2 XY gonads (XY FOXL2+ hereafter) by whole mount immunofluorescence for SF1 and FOXL2 (Fig. 2B–G). In contrast to the control XY gonads where FOXL2 was absent, FOXL2 protein was detected in almost all SF1+ cells in the XY FOXL2+ gonads at E11.5. At E14.5, Foxl2 mRNA was strongly expressed in XY FOXL2+ gonads and was about 4-folds the level of Foxl2 transcript in control XX gonads (Fig. 2H). We then investigated the effect of ectopic FOXL2 expression on testis differentiation. At both E14.5 and postnatal day 9 (P9), XY FOXL2+ gonads were smaller than control testes, and developed a shape similar to control ovaries (Fig. 2I–K); however, the male reproductive tract was maintained (Fig. 2L–M). Eight-week-old adult XY FOXL2+ mice presented a female-like external phenotype, with an anogenital distance similar to that of the control females (Fig. 2O–Q; Supplementary Material, Fig. S3A). The adult XY FOXL2+ mice produced significantly less testosterone than control males but they did not have an increase in estrogen production (Supplementary Material, Fig. S3B). Histologically, XY FOXL2+ adult gonads were dysgenic, forming a small structure that contained a few rudimentary seminiferous tubules in the center (Fig. 2R–T and Supplementary Material, Fig. S3C). No sign of spermatogenesis or folliculogenesis was detected in the XY FOXL2+ adult gonads. These results indicate that ectopic expression of FOXL2 in the SF1+ somatic cells of XY fetal gonads suppresses proper testis differentiation but is insufficient to induce a complete testis-to-ovary sex reversal.
FOXL2 represses Sertoli cell differentiation
Differentiation of gonadal primordium into a testis or an ovary relies on the capacity of the supporting cell population to become Sertoli cells or granulosa cells, respectively. We therefore focused on the fate of the supporting cells in XY FOXL2+ fetal gonads. At E11.5, the master testis-determining gene SRY and SOX9 were expressed in an XY-specific manner in the control gonads (Fig. 3A–C and G–I). At this early stage of fate determination, FOXL2 was absent in the XY control gonads and barely detected in the XX control gonads (Fig. 3D–F) as expected (6). Despite a proper expression of SRY (Fig. 3B and Supplementary Material, Fig. S3D), fewer SOX9+ Sertoli cells were found in the XY FOXL+ gonads at E11.5 compared to the control (Fig. 3G, H, K and L). These SOX9+ cells in XY FOXL2+ gonads co-expressed FOXL2 throughout fetal development (inset in Fig. 3E, H, L, O and R). In XY control gonads, SOX9+ Sertoli cell population rapidly expanded and formed testis cords. However, in XY FOXL2+ gonads, SOX9+ cell population remained relatively small and failed to form testis cords at E12.5 (Fig. 3N–S). SOX9+ cells were mostly located in the center of the XY FOXL2+ gonads. Labelling with the Sertoli cells cytoplasmic marker AMH showed that these cells eventually organized to form a few testis cords by E16.5 (arrows in Fig. 3U). SOX9 expression in the XY FOXL2+ gonads was further repressed postnatally (Supplementary Material, Fig. S3C). These results demonstrate that ectopic FOXL2 expression in SF1+ somatic cells of XY gonads represses Sertoli cell differentiation at the onset of testis determination.
Figure 3.
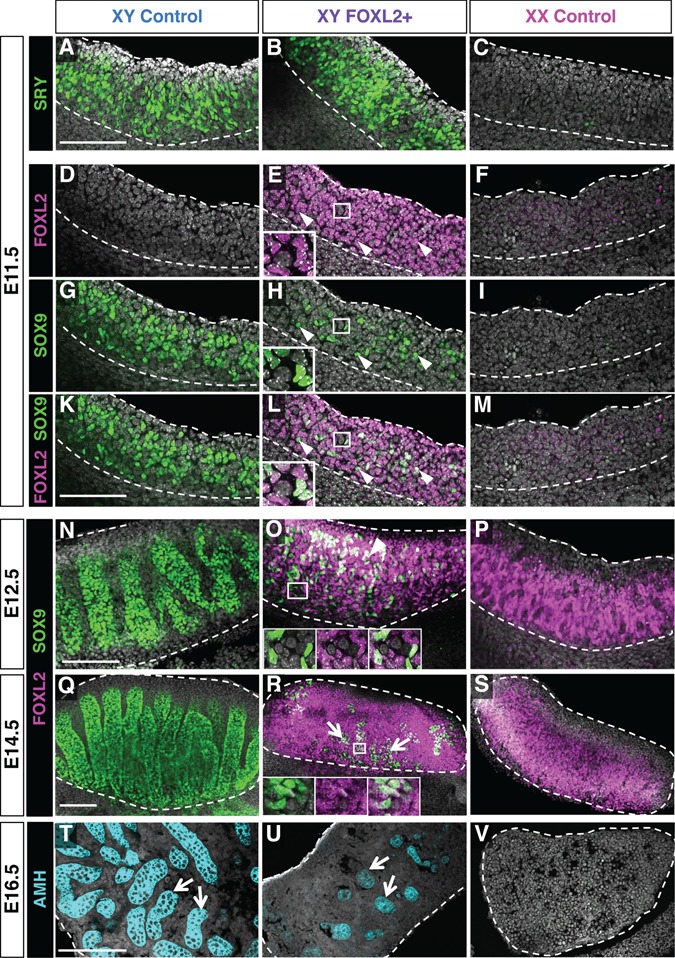
Impact of ectopic FOXL2 expression on Sertoli cell differentiation. Whole mount immunofluorescences for the testis-determining gene SRY at E11.5 (A–C), Sertoli cell marker SOX9 and FOXL2 at E11.5 (D–M), E12.5 (N–P) and E14.5 (Q–S) in XY control, XY FOXL2+ and XX control gonads. Arrowheads: cells double positive for SOX9 and FOXL2. Arrows in (R): SOX9+ cells are mainly located at the center of the gonads. Insets: higher magnification images for single channels showing the presence of FOXL2 in the SOX9+ cells. (T–V) Immunofluorescence for the Sertoli cell marker Anti-Müllerian hormone (AMH) on sections of E16.5 gonads showing the appearance of a few testis tubules in the center of XY FOXL2+ gonads (arrows). Dotted lines outline the gonads. Nuclei were stained with DAPI (gray). Scale bars: 150 μm.
Identification of the supporting cell genes affected by ectopic FOXL2 induction in XY gonads
To decipher the molecular events downstream of FOXL2 that led to defects in supporting cells differentiation (Fig. 3) and partial male-to-female sex reversal (Fig. 2), we performed microarray analyses on XY control, XY FOXL2+ and XX control gonads at E14.5 [Supplementary Material, Datasets S3–4; Gene Expression Omnibus (GEO) accession no. GSE78774]. Ectopic FOXL2 expression in the XY gonad shifted the gonadal transcriptome toward the ovarian profile, suggesting a partial testis-to-ovary sex-reversal (Fig. 4A). Between the XY FOXL2+ and XY control gonads, 1862 transcripts were significantly different, among them 1089 genes down-regulated and 755 genes upregulated (Fig. 4B; Supplementary Materials, Fig. S4 and Dataset S4). Most of the transcripts that were downregulated (81%) corresponded to transcripts normally expressed higher in the fetal testis than ovary. Whereas most of the transcripts that were upregulated (73%) corresponded to transcripts that were enriched in the fetal ovary compared to the testis (Supplementary Material, Fig. S4A and B). To identify the granulosa cell-specific genes upregulated as a consequence of FOXL2 induction, we compared the 755 upregulated genes with the 1179 genes enriched in granulosa cells versus Sertoli cells based on Jameson et al. (18) microarray (Fig. 4C). Similarly, we compared the 1089 downregulated genes with the 1410 genes enriched in Sertoli cells based on Jameson et al. microarray in order to identify the Sertoli cell genes significantly repressed in XY FOXL2+ gonads (Fig. 4D). These comparisons revealed 195 granulosa cell genes and 295 Sertoli cell genes that were significantly affected by ectopic FOXL2 induction in the fetal testis. Among the 195 granulosa-cell genes upregulated in XY FOXL2+ gonads, (GO) term analysis for biological processes revealed enrichments for genes associated with the Wnt pathway, a key pathway for ovarian differentiation (26,27; Fig. 4E and F; Supplementary Material, Dataset S4). Interestingly, GO terms associated with ‘negative regulation of cell cycle’ were also enriched, with upregulation of both the cyclin-dependent protein kinases Cdkn1b and Cdkn1c. These two genes are suspected to contribute to granulosa-cell exit of cell cycle and to the smaller size of ovaries compared to testes (28,29). In addition, Cdkn1b and Cdkn1c are strong candidates for a direct regulation by FOXL2 because they are known to be controlled by several other forkhead factors, such as FOXL1 and FOXOs (30,31). Several genes associated with ovarian differentiation including Bmp2 and Runx1 were also upregulated in XY FOXL2+ gonads (Fig. 4E and F). However, despite the high expression of Foxl2 in the XY FOXL2+ gonads (Fig. 2H), the expression of most granulosa-cell genes never reached the level of the control XX gonads (Fig. 4F). On the other hand, among the 295 Sertoli-cell genes downregulated in XY FOXL2+ gonads, many key Sertoli cell markers important for testis differentiation were identified, including Sox9, Amh, Ptgds, Amh, Gdnf and Cyp26b1 (Fig. 4E and F). These results imply that FOXL2 is a strong repressor of Sertoli cell differentiation and testis morphogenesis, but not sufficient to fully promote granulosa cell differentiation.
Figure 4.
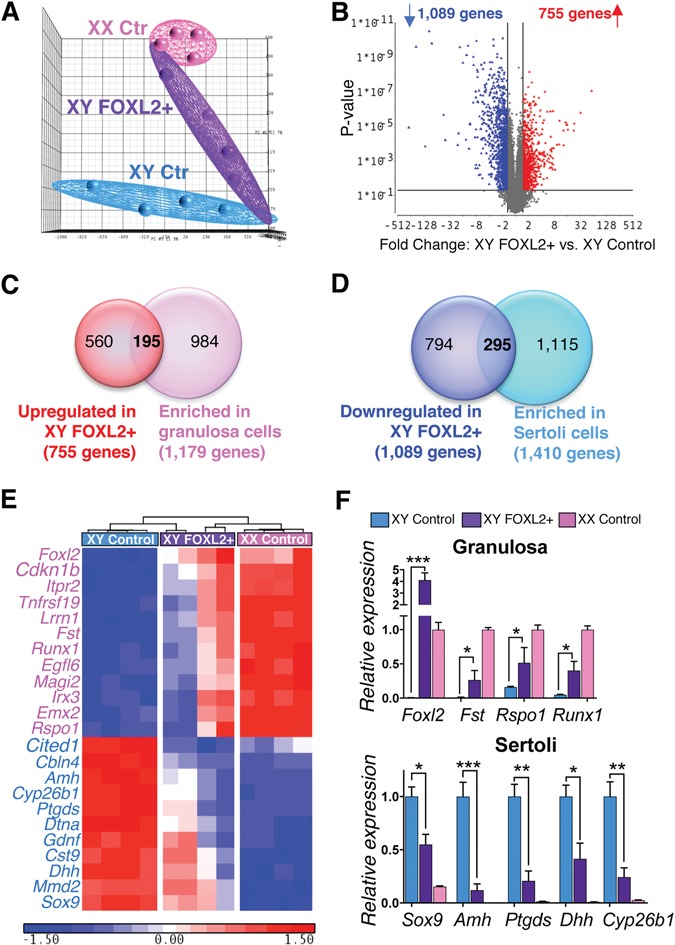
Identification of the genes affected by ectopic FOXL2 expression in the XY fetal gonads. (A) Principal component analysis for the transcriptomes of E14.5 XY control, XY FOXL2+ and XX control gonads (n = 4/genotype). (B) Volcano plot comparing transcript expression between E14.5 XY FOXL2+ and XY control gonads. Each dot represents a transcript; x-axis: fold-change expression; y-axis: P-value. Blue and red dots: transcripts significantly downregulated and upregulated, respectively (fold-change > 1.5; p0.05). (C) Venn diagram comparing the 755 genes upregulated in XY FOXL2 gonads with genes enriched in fetal granulosa cells versus Sertoli cells (based on (18); fold-change > 1.5 p0.05). (D) Venn diagram comparing the 1089 genes downregulated in XY FOXL2 gonads with genes enriched in fetal Sertoli cells versus granulosa cells (based on (18); fold-change > 1.5 p0.05). (E) Hierarchical clustering of a subset of granulosa cell genes upregulated (pink font) or Sertoli cell genes downregulated (blue font) in XY FOXL2+ gonads based on the comparison in C and D. (F) Validation of the microarray results by qPCR for genes identified in E. t-test; mean ± SEM (n = 4); *P < 0.05, **P < 0.01, ***P < 0.001.
Once the supporting cells (Sertoli or granulosa cells) are established, they control the differentiation of other cell populations in the fetal gonads. As a consequence of the repression of Sertoli cell differentiation and emergence of the granulosa cell program, both germ cells and steroidogenic precursors followed the feminizing trend of the supporting cells. XY FOXL2+ gonads expressed meiosis-related and oocyte-specific genes whereas fetal male germ cell markers were repressed (Supplementary Material, Fig. S4C). In addition, the androgen-producing Leydig cells, characteristic of fetal testis differentiation, failed to gain full steroidogenic capacity in XY FOXL2+ gonads, as evident by a decrease or absence of steroidogenic gene expression (Supplementary Material, Fig. S4D). Meiotic germ cells in the XY FOXL2+ gonads were located at the poles whereas the remnant features of testis differentiation (SOX9+ Sertoli cells and CYP17A1+Leydig cells) were found in the center (Supplementary Material, Fig. S4E–J), forming the typical structure of XY ovotestes (32–35).
Integrated analysis of transcriptomes of Foxl2 loss- and gain-of-function models with FOXL2 ChIP-seq data identifies strong candidates for direct regulation by FOXL2 during gonad differentiation
The transcriptomic analyses of the XY FOXL2+ gonads and the XX Foxl2 KO gonads provide complementary models to understand how FOXL2 contributes to supporting cell fate and gonad differentiation. Similar to the analysis for Foxl2 KO gonads (Fig. 1E and F), the genes differentially expressed in XY FOXL2+ gonads were compared with our FOXL2 ChIP-seq data (Supplementary Materials, Fig. S5 and Dataset S5). Identification of the genes commonly misregulated in both XY FOXL2+ and XX Foxl2 KO gonads allows us to focus on the genes that strongly rely on FOXL2 action, either negatively or positively (Fig. 5; Supplementary Material, Dataset S5). First, by comparing the genes significantly upregulated in XY FOXL2+ with the genes downregulated in XX Foxl2 KO gonads (Fig. 5A), 200 common genes were identified as genes stimulated by FOXL2. The majority of them (140 genes or 70%; Fig. 5A) corresponded to potential FOXL2 direct targets based on the FOXL2 ChIP-seq in the fetal ovary. Among these 140 genes, 62 genes were enriched in fetal granulosa cells, including Fst, the cell-cycle repressor Cdkn1b and several genes whose ovarian function is currently unknown, such as Hmgcs2, Magi2 and Itpr2. On the other hand, 121 genes were identified as genes suppressed by FOXL2 by comparing the genes downregulated in XY FOXL2+ gonads with the genes upregulated in XX Foxl2 KO gonad (Fig. 5B). Among them, 56 genes (46%) were identified in the FOXL2 ChIP-seq and included 26 genes enriched in fetal Sertoli cells, including Cyp26b1, Hsd17b3, Cldn11 and Erbb4. Interestingly, the testis-determining gene Sox9 was part of the 121 genes affected in both models (Supplementary Material, Dataset S5). However, similar to the FOXL2 ChIP-seq performed in adult granulosa cells (14), no FOXL2 binding sites were found near Sox9 gene in the fetal ovary, including in the testis-specific enhancer TES (Supplementary Material, Fig. S6). The nearest FOXL2 binding peak upstream of Sox9 was located in a gene desert 838 kb from Sox9 gene. The same region was also bound by FOXL2 in adult granulosa cells (14) and is conserved in many vertebrate species (Supplementary Material, Fig. S6). Interestingly, this region corresponds to a newly identified long-distance putative regulatory element for Sox9 (36). In the XY FOXL2+ gonads, downregulation of Sox9 was observed at both the transcript and the protein level (Figs 3 and 4), and likely contributed to the partial testis-to-ovary sex-reversal phenotype.
Figure 5.
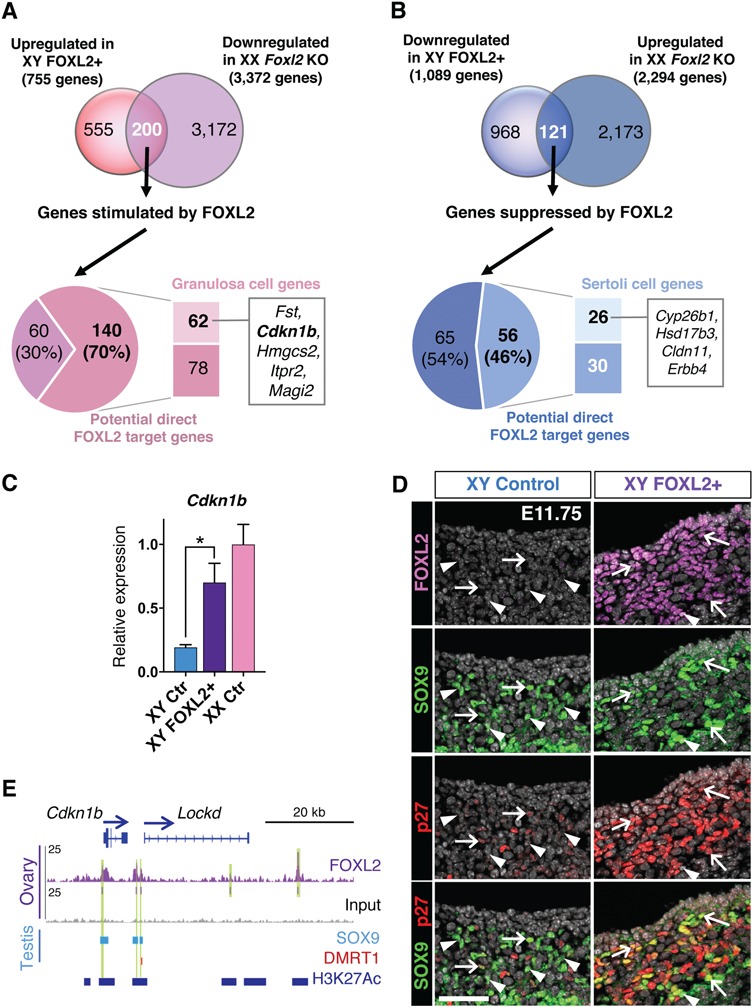
Comparison of XY FOXL2+ and XX Foxl2 KO transcriptomes with FOXL2 ChIP-seq gene list reveals strong candidates for direct regulation by FOXL2. (A) Comparison of the genes upregulated in XY FOXL2+ gonads and downregulated in XX Foxl2 KO gonads (13). Among the 200 common genes affected in both genetic models, 70% are potential direct targets of FOXL2 based on the ChIP-seq results and include 62 genes that are enriched in granulosa cells (18). (B) Comparison of the genes downregulated in XY FOXL2+ gonads and upregulated in XX Foxl2 KO gonads. Among the 121 genes affected in both models, 46% are potential direct targets of FOXL2 based on the ChIP-seq results and include 26 genes that are enriched in Sertoli cells. (C–E) Example of a potential direct target gene of FOXL2 enriched in granulosa cells: the cell-cycle repressor gene Cdkn1b. (C) qPCR analyses of Cdkn1b expression in E14.5 XY control, XY FOXL2+ and XX control gonads. t-test; mean ± SEM (n = 4); *P < 0.05, ***P < 0.001. (D) Immunofluorescence for FOXL2, SOX9 and p27 (encoded by Cdkn1b) in XY control and FOXL2+ gonads at E11.75. Arrows: SOX9+/p27+ double positive cells; Arrowheads: SOX9+/p27- cells. (E) Genome browser view of FOXL2-bound regions near Cdkn1b. SOX9 and DMRT1 tracks: genomic regions significantly bound by SOX9 (42) and DMRT1 (41) in E13.5 testis. H3K27Ac track: regions significantly bound by H3K27Ac in fetal Sertoli cells, corresponding to active chromatin regions (20). Arrows: gene orientation; asterisks: consensus FOXL2 motif. Green highlights: significant FOXL2 binding peaks.
Among the genes commonly affected in FOXL2 gain- and loss-of-function models, the cell-cycle repressor Cdkn1b was one of the most upregulated granulosa cell gene in XY FOXL2+ gonads (Fig. 5C). Because of its involvement in the control of cell proliferation, Cdkn1b was a functionally relevant candidate. Indeed, difference in cell proliferation is a sexually dimorphic event during sex determination: from E11.5–E13.5, the testis doubles its size every 24 h, while the growth of the ovary remains limited (37–39). After some recruitment from the coelomic epithelium and migration into the gonad, Sertoli cells resume proliferation while granulosa cells remain quiescent (28,38). The smaller SOX9+ Sertoli cell population in the XY FOXL2+ gonads (Fig. 3) prompted us to examine the expression of p27 (protein encoded by Cdkn1b) and the cell proliferation marker KI67. At E11.75 (21–23 tail somite stage), p27 was scarcely detected in few of SOX9+ Sertoli cells in the control XY gonads whereas KI67 was found in multiple SOX9+ cells at E11.75 and E12.5 (Fig. 5D; Supplementary Material, S7). In the FOXL2+ XY gonads, on the other hand, p27 was present in most SOX9+ supporting cells whereas only few of SOX9+ cells were KI67 positive (Fig. 5D; Supplementary Material, S7). These results suggest that upregulation of Cdkn1b, a potential direct target of FOXL2, could contribute to the smaller SOX9+ cell population by repressing their proliferation.
Several FOXL2-bound regions were located near Cdkn1b gene, including at its enhancer between the Cdkn1b gene body and the long non-coding RNA Lockd (Fig. 5E). This regulatory element plays a key role in the control of Cdkn1b expression as its deletion drastically reduces Cdkn1b expression (40). In the fetal Sertoli cells, this region is bound by the active chromatin marker H3K27Ac (Fig. 5E; 20), suggesting that FOXL2 could have access to these regions in XY FOXL2+ gonads in order to control Cdkn1b expression. Interestingly, this enhancer is also bound by SOX9 and DMRT1 based on ChIP-seq performed in fetal testes (Fig. 5E; 41,42). These observations together suggest that Cdkn1b is controlled by both pro-ovarian FOXL2 and pro-testis transcription factors during gonad differentiation.
Pro-granulosa FOXL2 and pro-Sertoli regulator SOX9 share common chromatin binding regions in fetal gonads
SOX9 and DMRT1 are fate determining/maintenance factors for Sertoli cell lineage, and pro-granulosa FOXL2 is known to antagonize these two factors (10,43). The discovery of the presence of FOXL2, SOX9 and DMRT1 binding in the enhancer region of Cdkn1b (Fig. 5E) and other potential FOXL2 direct target genes expressed in granulosa or Sertoli cells (Supplementary Material, Fig. S5C) raised the question whether these three transcription factors could bind similar regulatory regions in more genes in the fetal gonads. We therefore compared our genome-wide FOXL2 ChIP-seq results in the fetal ovary with published ChIP-seq results for SOX9 and DMRT1 in the fetal testis (41,42; Fig. 6; Supplementary Material, Dataset S6). Among the 11 438 FOXL2 binding peaks from our ChIP-seq analysis (Fig. 6A), 35% (4008 peaks) overlapped with SOX9 peaks only, 3% (290 peaks) with DMRT1 peaks only and 2% (257 peaks) with both SOX9 and DMRT1 peaks (Fig. 6A). Overall, 62% of SOX9 binding peaks overlapped with FOXL2 peaks (Fig. 6B; 3980/6400 peaks) whereas only 23% of DMRT1 peaks overlapped with FOXL2 peaks (Fig. 6C; 563/2458 peaks). These results demonstrate an extensive overlap between FOXL2 and SOX9 chromatin occupancy, and suggest that FOXL2 and SOX9 could share common target genes during gonad differentiation. Some of these chromatin regions bound by both FOXL2 and SOX9 were located at the promoter of genes either upregulated or downregulated in XY FOXL2+ gonads (Supplementary Material, Dataset S6). For instance, Cited2, a gene that contributes to early differentiation of the testis (44), was significantly downregulated in XY FOXL2+ gonads and was identified as a common target of FOXL2 and SOX9 (Fig. 6D). Very few genes that were affected in XY FOXL2+ gonads contained chromatin regions with overlapping binding of FOXL2 and DMRT1. Among them was Notch2, a gene downregulated in XY FOXL2+ gonads, whose promoter was bound by FOXL2, DMRT1 and SOX9 in fetal gonads (Fig. 6D).
Figure 6.
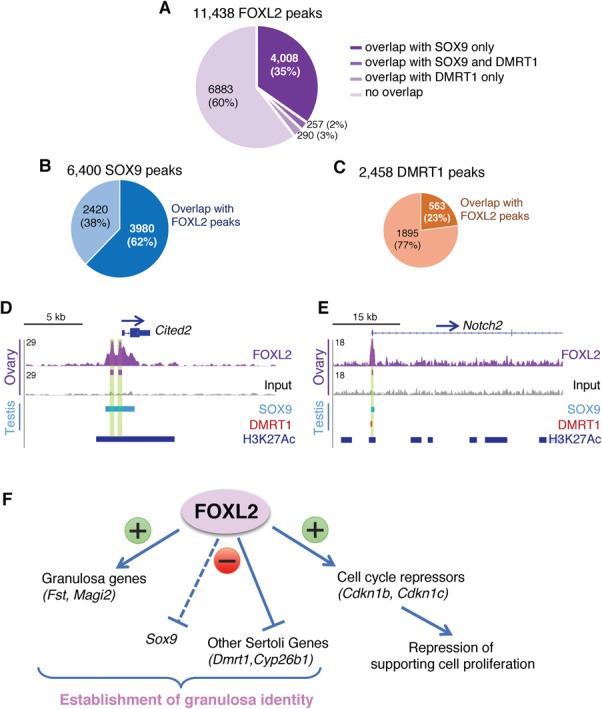
Comparison of FOXL2 genome occupancy in the fetal ovary with SOX9 and DMRT1 genome occupancy in the fetal testis reveals extensive overlaps between FOXL2 and SOX9 binding regions. (A) Distribution of FOXL2 peaks based on their overlap with SOX9 or/and DMRT1 peaks. (B) Percentage of SOX9 peaks that overlap with FOXL2 peaks. (C) Percentage of DMRT1 peaks that overlap with FOXL2 peaks. (D–E) Genome browser view of Cited2 and Notch2 genes bound by FOXL2, SOX9 and/or DMRT1. SOX9 and DMRT1 tracks: genomic regions significantly bound by SOX9 (42) and DMRT1 (41) in E13.5 testis. H3K27Ac track: regions significantly bound by H3K27Ac in fetal Sertoli cells, corresponding to active chromatin regions (20). Arrows: gene orientation; asterisks: consensus FOXL2 motif. Green highlights: significant FOXL2 binding peaks. (F) Summary of FOXL2 functions in the fetal gonads based on FOXL2 ChIP-seq and loss- and gain-of-function mouse models. FOXL2 contributes to granulosa cell identity through direct stimulation of granulosa cell-specific genes and suppression of Sertoli cell-specific genes. Multiple targets of FOXL2 are linked to control of cell cycle, potentially contributing to the cell cycle arrest of fetal granulosa cells.
Discussion
Sex differentiation of the gonads is regulated by opposing networks of transcription factors that promote one fate (testis or ovary) while repressing the other. In the female embryo, ovarian differentiation relies on the action of two major regulators: the RSPO1/WNT4 beta-catenin pathway and FOXL2. To determine how FOXL2 controls the fate of supporting cells in the fetal gonads, we first performed ChIP-seq for FOXL2 in fetal ovaries and identified its potential target genes. We next generated a mouse model that allows ectopic expression of FOXL2 specifically in somatic cells of XY gonads at the time of sex determination. This model showed that presence of FOXL2 in the supporting cells of fetal testes repressed early Sertoli cell differentiation and resulted in partial testis-to-ovary sex reversal. The transcriptome analysis of this new model produced a list of genes and pathways affected by FOXL2 induction. Comparison of this gain-of-function model with the published Foxl2 KO model and our ChIP-seq results yielded a list of biologically relevant FOXL2 targets. Positive regulation of genes enriched in granulosa cells and negative regulation of genes enriched in Sertoli cells could tip the balance toward the ovarian pathway (Fig. 6F). In addition, a strong correlation between FOXL2 and the control of cell cycle further supports a role of FOXL2 in mitotic arrest of granulosa cells. Finally, by comparing our FOXL2 ChIP-seq data in the fetal ovary with previously published ChIP-seq for SOX9 and DMRT1 in the fetal testis, we discovered an extensive overlap in chromatin occupancy between FOXL2 and SOX9, suggesting that they control common regulatory regions during gonad differentiation.
Deciphering FOXL2 functions in fate determination: lessons learned from the gain-of-function mouse model
Ectopic FOXL2 expression in SF1+ somatic cells of fetal XY gonads antagonizes Sertoli cell differentiation and testis morphogenesis, and results in partial testis-to-ovary sex reversal. Following ectopic FOXL2 induction, expression of the testis-determining factor SOX9 is decreased and remains mostly restricted to the center of the gonad. Sox9 is a key player for testis differentiation: its expression is sufficient to induce ovary-to-testis sex reversal (45) and its loss leads to testis-to-ovary sex reversal (46). The decrease, but not absence, of SOX9 expression could explain the partial testis-to-ovary sex reversal in XY FOXL2+ gonads. On the other hand, while Sox9 transcript is detected in XX Foxl2 KO fetal gonads (13), its protein is only expressed after birth and is likely the reason why the ovary-to-testis sex-reversal only occurs postnatally in absence of Foxl2 (9).
It remains unclear whether FOXL2 directly inhibits Sox9 transcription in the fetal gonad. In adult granulosa cells, FOXL2, along with estrogen signaling, represses Sox9 expression via occupying the Sox9-regulatory sequence TES (10). Yet, FOXL2 ChIP-seq experiments on fetal ovaries (this study) and adult granulosa cells (14) did not detect any FOXL2 binding at the TES. In XY mice, removal of TES only caused a 50% reduction of Sox9 expression without the occurrence of testis-to-ovary sex reversal, suggesting the existence of additional enhancers that control Sox9 expression in the fetal testis (36). Regulation of SOX9 expression is complex, and multiple tissue-specific enhancer regions have been identified as far as 2 Mb from SOX9 gene in human (47). Recently, several novel gonadal regulatory elements were identified upstream of Sox9 gene in the mouse genome, including enhancer 13 (Enh13), an essential regulatory element for initiation of testis development (36). Another putative long-distance regulatory element, Enh8, is located in an open chromatin region in fetal granulosa cells and is suspected to be required to repress Sox9 gene in the granulosa cells. Interestingly, in both fetal ovaries (this study) and in adult granulosa cells (14), Enh8 is bound by FOXL2. It is therefore possible that FOXL2 acts through this long-distance regulatory element region to repress Sox9 expression in both fetal gonads and adult granulosa cells. Another potential mechanism is that FOXL2 represses Sox9 expression indirectly through its capacity to interact with SF1 (48,49). Since SF1 is a co-factor of both SRY and SOX9 (50), interaction between FOXL2 and SF1 could disrupt SF1 availability to promote Sox9 expression, and could affect SOX9 transcriptomic function itself.
In XY FOXL2+ gonads, initial SOX9 expression is not completely prevented, and this is likely the cause of the incomplete testis-to-ovary sex reversal in XY FOXL2+ gonads. We speculate that the timing of FOXL2 induction in our model, which is slightly after SRY expression is initiated (51), could prevent a complete repression of SOX9. Ectopic presence of FOXL2 did not affect expression of SRY, which is known to induce Sox9 expression. Among the potential FOXL2 direct target genes, some are genes involved in controlling SRY expression, such as Cbx2, or SOX9 expression, such as Cited2 (44,52,53). We wonder whether an earlier induction of FOXL2 could fully prevent SOX9 expression and even affect SRY expression. Unfortunately, attempts to induce FOXL2 earlier using another Cre model (Wt1CreERT2) resulted in early embryonic lethality, likely due to the impact of ectopic FOXL2 expression in other WT1+ organs such as the heart.
Another possibility is that FOXL2 alone is insufficient to fully repress Sox9 expression in the fetal XY gonads. The population of SOX9+ cells in the XY FOXL2+ gonad is positive for FOXL2 and is able to maintain SOX9 expression throughout fetal development despite the presence of FOXL2. These SOX9+ cells in the XY FOXL2+ gonads are located at the center of the gonads, similar to other testis-to-ovary sex reversal models (32–35). The supporting cells at the center of XY gonads are the first to express SRY (54), possibly making them more committed to the testis fate and more resistant to the feminizing effect of FOXL2.
FOXL2, a regulator of somatic cell proliferation in the gonads
In the XY FOXL2+ gonads, the population of SOX9+ Sertoli cells was significantly reduced, suggesting that the presence of ectopic FOXL2 interferes with the expansion of Sertoli cell population. One characteristic difference between fetal Sertoli cells and granulosa cells is their proliferative status: granulosa cells in XX gonads are p27+ and mitotically arrested (28,55) whereas Sertoli cells in XY fetal gonads become p27- and proliferative (28,38). Following FOXL2 ectopic expression, supporting cells in the XY gonad become p27+ and fail to proliferate, suggesting that they acquired a cell cycle status similar to fetal granulosa cells. It has been shown that overexpression of Cdkn1b (the gene that encodes p27) is sufficient to repress cell proliferation in vivo (56). FOXL2, similar to other forkhead transcription factors, is associated with repression of cell proliferation (57–59). In addition to Cdkn1b, FOXL2 ChIP-seq analysis in the fetal ovary as well as XY FOXL2+ transcriptome reveals that multiple genes/pathways involved in cell-cycle control are potential targets of FOXL2, such as Cdkn1c, another cell cycle repressor gene enriched in granulosa cells (29). These genes could also contribute to the repression of Sertoli cell proliferation in XY FOXL2+ gonads, and to the mitotic arrest status of fetal granulosa cells during normal ovarian development. Interestingly, characterization of other ovary-to-testis sex-reversal models suggests that exit of the mitotic arrest of pre-granulosa cells is a prerequisite for Sox9 upregulation in these models (60). Although loss of Wnt4 or Foxl2 affects Cdkn1b expression during fetal life, it does not lead to a complete repression and fails to disrupt fetal granulosa cell mitotic arrest, likely due to compensation by other ovarian signals. As a consequence, loss of Wnt4 or Foxl2 results in SOX9 expression only in neonatal/postnatal ovary, when granulosa cell proliferation is normally initiated (7,27). Based on this hypothesis of interdependence between cell cycle and supporting cell fate (60), activation of cell cycle arrest in supporting cells of XY FOXL2+ gonads could also contribute to the defects in Sox9 expression in our gain-of-function model.
Toward the identification of chromatin regulatory regions controlling supporting cell differentiation
Fate specification of gonadal somatic cells relies on complex regulatory networks composed of multiple transcription factors. ChIP-seq has been a key analytical tool to understand the functions of a transcription factor in a tissue of interest. However, it remains challenging to apply this technique to fetal organs such as gonads due to the limited amount of tissues (hundreds of gonads for one ChIP-seq for a transcription factor) and the complex cellular composition of the organs. To date, ChIP-seq was performed in fetal gonads for only two other transcription factors: SOX9 and DMRT1 (41,42), both critical for Sertoli cell identity. By comparing the results from these three ChIP-seq experiments, we observed an extensive overlap between FOXL2 genome-wide occupancy in the fetal ovary and SOX9 occupancy in the fetal testis. These common regulatory elements could be the targets for both gene activation and gene repression depending on the transcription factors bound during gonad differentiation. The capacity of FOXL2 to bind genomic regions that are also bound by SOX9 could contribute to its capacity to antagonize Sertoli cell differentiation in XY FOXL2+ gonads. FOXL2 could compete with SOX9 for chromatin accessibility and therefore repress SOX9 activity.
An alternative to ChIP-seq for specific transcription factors is to identify the open chromatin domains during supporting cell differentiation. Maatouk et al. examined the chromatin status in fetal Sertoli cells by DNaseI-seq and H3K27Ac ChIP-seq, and identified regulatory sites contributing to either activation or repression of genes during Sertoli cell differentiation (20). Interestingly, their study showed that forkhead motifs were significantly enriched in active chromatin regions in addition to SOX motifs, consistent with our discovery of the overlapping chromatin occupancy for FOXL2 and SOX9 in fetal gonads.
In conclusion, our data provide insights into FOXL2 chromatin occupancy during ovarian differentiation and enable us to identify biologically relevant candidates downstream of FOXL2 action during gonad differentiation. The capacity of FOXL2 to bind similar chromatin regions with SOX9 suggests that some regulatory elements in the fetal gonads are targets of both pro-ovary and pro-testis transcription factors.
Materials and Methods
ChIP-seq assays and analysis
Ovaries from E14.5 CD1 embryos were separated from the mesonephros, snap-frozen and stored at −80°C. Two independent ChIP-seq experiments were performed by Active Motif Inc. (Carlsbad, CA) using 20–30 μg of sheared chromatin from pooled embryonic ovaries (n = 100–120 ovaries/ChIP), and 4 μg of FOXL2 antibody (Novus, NB100–1277, Littleton, CO). ChIP-seq libraries were sequenced as single-end 75-mers by Illumina NextSeq 500, then filtered to retain only reads with average base quality score > 20. Reads were mapped against the mouse mm10 reference genome using Bowtie v1.2 (61) with parameter ‘-m 1’ to collect only uniquely-mapped hits. Duplicate mapped reads were removed using Picard tools MarkDuplicates.jar (v1.110). The number of uniquely-mapped non-duplicate reads for each biological replicate was 18 509 700 and 15 249 438. After merging the replicate datasets, binding regions were identified by peak calling using HOMER v.4.9 (62) with FDR < 1e-5. Called peaks were subsequently redefined as 300mers centered on the called peak midpoints. The peaks were filtered for a required 4-fold enrichment over input and over local signal. Genomic distribution of FOXL2-bound regions was determined based on Refseq gene models as downloaded from the UCSC Genome Browser as of February 27, 2017. Enriched motifs were identified using HOMER findMotifsGenome.pl de novo motif analysis with parameter ‘-size given’. Peaks were assigned to the nearest gene (63), again based on RefSeq gene models. Gene lists were analyzed for enrichment using the online tool EnrichR (15). The ChIP-seq data are available in GEO (GSE110093; http://www.ncbi.nlm.nih.gov/geo/). To compare FOXL2-bound regions in fetal ovary to SOX9 and DMRT1-bound regions in the fetal testis (41,42), published peak sets for SOX9 and DMRT1-bound chromatin regions were converted to mouse mm10 genome using UCSC LiftOver tool. This conversion resulted in total usable counts of 6400 peaks for SOX9 and 2458 peaks for DMRT1. Gene comparisons were performed using Partek and the online tool Venny 2.1 (http://bioinfogp.cnb.csic.es/tools/venny/index.html).
Generation of Rosa26-CAG-LSL-Foxl2 mice
Foxl2 coding sequence with a HIS-tag on 3′ of the sequence was amplified from a mouse BAC (# 231A10, Invitrogen) using the primers Foxl2_ORF_Fw 5′-CGGCCGGCCGCCACCATGATGGCCAGCTACCCCGAGC-3′ and Foxl2-His-Cterm_Rv 5′-CGGCCGGCCTCAGTGGTGATGGTGATGATGGAGATCCAGACGCGAGTGCA-3′. This Foxl2-HisTag sequence was inserted into the pAi9 vector (64) (Addgene) in substitution of the dTomato sequence using FseI restriction sites. Embryonic stem (ES) cells (129SvEv) were electroporated with this modified vector in order to insert the targeting construct into Rosa26 locus by homologous recombination. Targeted 129SvEv ES cells were identified by Southern blot and injected into C57-BL6 albino blastocysts to generate the founder mice. A founder male was mated to C57BL/6 J females (Supplementary Material, Fig. S1C) and the resulting mouse line was maintained in the same mixed genetic background (129Sv and C57Bl/6 J). Genotyping of the animals was performed by PCR using the Rosa-Ctr_Fw 5′-AAGGGAGCTGCAGTGGAGTA-3′; Rosa-Ctr_Rv 5′-CCGAAAATCTGTGGGAAGTC-3′ primers for the wild-type allele (297 bp band size), and Rosa-Foxl2_Fw 5′-CTCATACTGGGACCACGACA-3′; Rosa-Foxl2_Rv 5′-GGCATTAAAGCAGCGTATCC-3′ primers for the flox allele (209 bp band size). The PCR analyses included 35 cycles of reaction at the following temperature: 30 sec at 95°C, 30 sec at 61°C, 30 sec at 72°C, followed by 3 min of terminal elongation at 72°C.
Mouse model for ectopic induction of FOXL2 expression
For ectopic induction of FOXL2 expression in fetal gonads, Rosa26-CAG-LSL-Foxl2 mice were time-mated overnight with Sf1-cre mice (21). Noon of the day when the vaginal plug was detected was considered as embryonic day E0.5. The Cre recombinase expression under the control of steroidogenic factor 1 (Sf1) regulatory elements allowed the conditional expression of FOXL2 in the somatic cells of undifferentiated gonads. The genotypes of XY FOXL2+ and their control littermates are Rosa26-CAG-LSL-Foxl2+/f; Sf1-CreTg/+ and Rosa26+/+; Sf1-CreTg/+ or Rosa26-CAG-LSL-Foxl2+/f, respectively. All animal procedures were approved by the National Institute of Environmental Health Sciences (NIEHS) Animal Care and Use Committee and are in compliance with a NIEHS-approved animal study proposal.
Immunofluorescence and histological analysis
Gonads were collected and fixed overnight at 4°C in 4% paraformaldehyde. The immunofluorescence experiments were performed on whole mount gonads at E11.5 (18–20 tail somite stage), E12.5 and E14.5, or on 8 μm cryosections for E11.75 (21–23 tail somites), E14.5 and E16.5 gonads, or on 7 μm paraffin sections for E12.5 and P9 gonads. For paraffin-embedded tissues, the sections were dewaxed and rehydrated in a decreasing gradient of alcohol. The slides were pretreated in 0.1 mm citric acid for 20 min in the microwave and cooled down at room temperature. All the samples (whole mount gonads, cryosections and paraffin sections) were blocked in PBS-Triton X-100 solution with 5% normal donkey serum for 1 h and incubated with primary antibodies at 4°C overnight. After washing in PBS-Triton X-100, the samples were incubated in the appropriate secondary antibody (1:500; Invitrogen, USA), then washed again and mounted in Vector Mount with DAPI (Vector Labs, Burlingame, CA). Whole mount gonads or sections were imaged under a Leica DMI4000 confocal microscope. Histological analysis was performed by staining dewaxed paraffin sections with hematoxylin and eosin. The stained sections were scanned using Aperio ScanScope XT Scanner (Aperio Technologies, Inc., CA, USA). Antibodies used in this study are listed in Supplementary Material, Table S1.
Real-time PCR analysis
Total RNA was isolated from gonad pairs using PicoPure RNA isolation kit (Arcturus, Mountain View, CA). 300 ng of RNA was used for cDNA synthesis with the Superscript II cDNA synthesis system (Invitrogen Corp., Carlsbad, CA). Gene expression was analyzed by real-time PCR using Bio-Rad CFX96TM Real-Time PCR Detection system. Gene expression was normalized to Gapdh. The Taqman probes and primers used to detect transcript expression are listed in Supplementary Material, Table S1. Four biological replicates were used and controls correspond to littermates of XY FOXL2+ mice lacking the Sf1-Cre transgene. Data were analyzed using Prism GraphPad Software by two-tailed Student’s t-test. Values are presented as mean ± S.E.M.
Microarray analysis
Gene expression analysis was conducted using Affymetrix Mouse Transcriptome 1.0 arrays (Affymetrix, Santa Clara, CA) on four biological replicates (one pair of E14.5 gonads) for each genotype. A total of 100 ng RNA were amplified and labeled using the Affymetrix WT Plus Reagent Kit (WT Plus Kit). A total of 5.5 μg micrograms of amplified biotin-cDNAs were fragmented and hybridized to each array for 16 h at 45°C in a rotating hybridization oven. Array slides were stained with streptavidin/phycoerythrin then washed for antibody amplification according to the GeneChip Hybridization, Wash and Stain Kit and user manual protocol FS450-0001. Arrays were scanned in an Affymetrix Scanner 3000 and data (GEO accession no. GSE78774) was obtained using the GeneChip® Command Console software. Gene-level normalization was conducted with Affymetrix Transcriptome Analysis Console 2.0 software and gene expression analyses were conducted with Partek software (St. Louis, Missouri). A full dataset Excel file containing the normalized log2 intensity of all genes for each genotype, and a graphic view of their expression are provided in Supplementary Material, Dataset S4. In order to identify differentially expressed genes, analysis of variance was used to determine if there was a statistical difference between the means of groups. The gene lists were filtered with fold-change cutoff of 1.5 and P < 0.05. Raw data from E13.5 granulosa and Sertoli cell transcriptomes (18) were reanalyzed using Partek in order to compare them to XY FOXL2+ transcriptome. Venn diagrams were generated in Partek by comparing gene symbols between the different microarray lists. Lists of genes differentially expressed in Foxl2 KO ovaries directly originate from published datasets on E13.5, E16.5 and newborn gonads (13).
Steroid hormone immunoassay
Serum samples were collected at the time of sacrifice and stored at −80°C until analyzed. Serum levels of estradiol and testosterone were measured by ELISA by the University of Virginia Center for Research in Reproduction Ligand Assay and Analysis Core.
Supplementary Material
Acknowledgements
We thank David Zarkower (University of Minnesota, USA) for the DMRT1 antibody, Ken Morohashi (Kyushu University, Japan) for the SF1 and SOX9 antibodies and Dagmar Wilhelm (University of Melbourne, Australia) for the SRY antibody. We are grateful for the NIEHS Molecular Genomics Core for the microarray analyses and Comparative Medicine Branch for mouse colony maintenance.
Conflict of Interest statement. None declared.
Funding
This work was supported by the Intramural Research Program (ES102965) of the NIH, National Institute of Environmental Health Sciences.
References
- 1. Herpin A. and Schartl M. (2015) Plasticity of gene-regulatory networks controlling sex determination: of masters, slaves, usual suspects, newcomers, and usurpators. EMBO Rep., 16, 1260–1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Crisponi L., Deiana M., Loi A., Chiappe F., Uda M.,Amati P., Bisceglia L., Zelante L., Nagaraja R., Porcu S. et al. (2001) The putative forkhead transcription factor FOXL2 is mutated in blepharophimosis/ptosis/epicanthus inversus syndrome. Nat. Genet., 27, 159–166. [DOI] [PubMed] [Google Scholar]
- 3. Bertho S., Pasquier J., Pan Q., Le Trionnaire G., Bobe J., Postlethwait J.H., Pailhoux E., Schartl M., Herpin A. and Guiguen Y. (2016) Foxl2 and its relatives are evolutionary conserved players in gonadal sex differentiation. Sex. Dev., 10, 111–129. [DOI] [PubMed] [Google Scholar]
- 4. Boulanger L., Pannetier M., Gall L., Allais-Bonnet A., Elzaiat M., Le Bourhis D., Daniel N., Richard C.,Cotinot C., Ghyselinck N.B. et al. (2014) FOXL2 is a female sex-determining gene in the goat. Curr. Biol., 24, 404–408. [DOI] [PubMed] [Google Scholar]
- 5. Li M., Yang H., Zhao J., Fang L., Shi H., Li M., Sun Y., Zhang X., Jiang D., Zhou L. et al. (2014) Efficient and heritable gene targeting in tilapia by CRISPR/Cas9. Genetics, 197, 591–599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Loffler K.A., Zarkower D. and Koopman P. (2003) Etiology of ovarian failure in blepharophimosis ptosis epicanthus inversus syndrome: FOXL2 is a conserved, early-acting gene in vertebrate ovarian development. Endocrinology, 144, 3237–3243. [DOI] [PubMed] [Google Scholar]
- 7. Schmidt D., Ovitt C.E., Anlag K., Fehsenfeld S., Gredsted L., Treier A.C. and Treier M. (2004) The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development, 131, 933–942. [DOI] [PubMed] [Google Scholar]
- 8. Uda M., Ottolenghi C., Deiana M., Kimber W., Forabosco A., Cao A., Schlessinger D. and Pilia G. (2004) Foxl2 disruption causes mouse ovarian failure by pervasive blockage offollicle development. Hum. Mol. Genet., 13, 1171–1181. [DOI] [PubMed] [Google Scholar]
- 9. Ottolenghi C., Omari S., Garcia-Ortiz J.E., Uda M., Crisponi L., Forabosco A., Pilia G. and Schlessinger D. (2005) Foxl2 is required for commitment to ovary differentiation. Hum. Mol. Genet., 14, 2053–2062. [DOI] [PubMed] [Google Scholar]
- 10. Uhlenhaut N.H., Jakob S., Anlag K., Eisenberger T., Sekido R., Kress J., Treier A.-C., Klugmann C., Klasen C., Holter N.I. et al. (2009) Somatic sex reprogramming of adult ovaries to testes by FOXL2 ablation. Cell, 139, 1130–1142. [DOI] [PubMed] [Google Scholar]
- 11. Auguste A., Chassot A.A., Gregoire E.P., Renault L., Pannetier M., Treier M., Pailhoux E. and Chaboissier M.C. (2011) Loss of R-spondin1 and Foxl2 amplifies female-to-male sex reversal in XX mice. Sex. Dev., 5, 304–317. [DOI] [PubMed] [Google Scholar]
- 12. Ottolenghi C., Pelosi E., Tran J., Colombino M., Douglass E., Nedorezov T., Cao A., Forabosco A. and Schlessinger D. (2007) Loss of Wnt4 and Foxl2 leads to female-to-male sex reversal extending to germ cells. Hum. Mol. Genet., 16, 2795–2804. [DOI] [PubMed] [Google Scholar]
- 13. Garcia-Ortiz J.E., Pelosi E., Omari S., Nedorezov T., Piao Y., Karmazin J., Uda M., Cao A., Cole S.W., Forabosco A. et al. (2009) Foxl2 functions in sex determination and histogenesis throughout mouse ovary development. BMC Dev.Biol., 9, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Georges A., L'Hote D., Todeschini A.L., Auguste A., Legois B., Zider A. and Veitia R.A. (2014) The transcription factor FOXL2 mobilizes estrogen signaling to maintain the identity of ovarian granulosa cells. Elife, 3, e04207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Chen E.Y., Tan C.M., Kou Y., Duan Q., Wang Z., Meirelles G.V., Clark N.R. and Ma'ayan A. (2013) Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics, 14, 128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kuleshov M.V., Jones M.R., Rouillard A.D., Fernandez N.F., Duan Q., Wang Z., Koplev S., Jenkins S.L., Jagodnik K.M., Lachmann A. et al. (2016) Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res., 44, W90–W97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Martins R., Lithgow G.J. and Link W. (2016) Long live FOXO: unraveling the role of FOXO proteins in aging and longevity. Aging Cell, 15, 196–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jameson S.A., Natarajan A., Cool J., DeFalco T., Maatouk D.M., Mork L., Munger S.C. and Capel B. (2012) Temporal transcriptional profiling of somatic and germ cells reveals biased lineage priming of sexual fate in the fetal mouse gonad. PLoS Genet., 8, e1002575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Blount A.L., Schmidt K., Justice N.J., Vale W.W., Fischer W.H. and Bilezikjian L.M. (2009) FoxL2 and Smad3 coordinately regulate follistatin gene transcription. J. Biol. Chem., 284, 7631–7645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Maatouk D.M., Natarajan A., Shibata Y., Song L., Crawford G.E., Ohler U. and Capel B. (2017) Genome-wide identification of regulatory elements in Sertoli cells. Development, 144, 720–730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Bingham N.C., Verma-Kurvari S., Parada L.F. and Parker K.L. (2006) Development of a steroidogenic factor 1/Cre transgenic mouse line. Genesis, 44, 419–424. [DOI] [PubMed] [Google Scholar]
- 22. Lavery R., Lardenois A., Ranc-Jianmotamedi F.,Pauper E., Gregoire E.P., Vigier C., Moreilhon C., Primig M. and Chaboissier M.C. (2011) XY Sox9 embryonic loss-of-function mouse mutants show complete sex reversal and produce partially fertile XY oocytes. Dev. Biol., 354, 111–122. [DOI] [PubMed] [Google Scholar]
- 23. Liu C.F., Bingham N., Parker K. and Yao H.H. (2009) Sex-specific roles of beta-catenin in mouse gonadal development. Hum. Mol. Genet., 18, 405–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Maatouk D.M., DiNapoli L., Alvers A., Parker K.L., Taketo M.M. and Capel B. (2008) Stabilization of beta-catenin in XY gonads causes male-to-female sex-reversal. Hum. Mol. Genet., 17, 2949–2955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Nicol B. and Yao H.H. (2015) Gonadal identity in the absence of pro-testis factor SOX9 and pro-ovary factor beta-catenin in mice. Biol. Reprod., 93, 35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Parma P., Radi O., Vidal V., Chaboissier M.C., Dellambra E., Valentini S., Guerra L., Schedl A. and Camerino G. (2006) R-spondin1 is essential in sex determination, skin differentiation and malignancy. Nat. Genet., 38, 1304–1309. [DOI] [PubMed] [Google Scholar]
- 27. Vainio S., Heikkila M., Kispert A., Chin N. and McMahon A.P. (1999) Female development in mammals is regulated by Wnt-4 signalling. Nature, 397, 405–409. [DOI] [PubMed] [Google Scholar]
- 28. Gustin S.E., Hogg K., Stringer J.M., Rastetter R.H., Pelosi E., Miles D.C., Sinclair A.H., Wilhelm D. and Western P.S. (2016) WNT/beta-catenin and p27/FOXL2 differentially regulate supporting cell proliferation in the developing ovary. Dev. Biol., 412, 250–260. [DOI] [PubMed] [Google Scholar]
- 29. Nef S., Schaad O., Stallings N.R., Cederroth C.R., Pitetti J.L., Schaer G., Malki S., Dubois-Dauphin M., Boizet-Bonhoure B., Descombes P. et al. (2005) Gene expression during sex determination reveals a robust female genetic program at the onset of ovarian development. Dev. Biol., 287, 361–377. [DOI] [PubMed] [Google Scholar]
- 30. Chen X., Deng M., Ma L., Zhou J., Xiao Y., Zhou X., Zhang C. and Wu M. (2015) Inhibitory effects of forkhead box L1 gene on osteosarcoma growth through the induction of cell cycle arrest and apoptosis. Oncol. Rep., 34, 265–271. [DOI] [PubMed] [Google Scholar]
- 31. Medema R.H., Kops G.J., Bos J.L. and Burgering B.M. (2000) AFX-like Forkhead transcription factors mediate cell-cycle regulation by Ras and PKB through p27kip1. Nature, 404, 782–787. [DOI] [PubMed] [Google Scholar]
- 32. Bagheri-Fam S., Sim H., Bernard P., Jayakody I., Taketo M.M., Scherer G. and Harley V.R. (2008) Loss of Fgfr2 leads to partial XY sex reversal. Dev. Biol., 314, 71–83. [DOI] [PubMed] [Google Scholar]
- 33. Bogani D., Siggers P., Brixey R., Warr N., Beddow S.,Edwards J., Williams D., Wilhelm D., Koopman P., Flavell R.A. et al. (2009) Loss of mitogen-activated protein kinase kinase kinase 4 (MAP3K4) reveals a requirement for MAPK signalling in mouse sex determination. PLoS Biol., 7, e1000196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Menke D.B., Koubova J. and Page D.C. (2003) Sexual differentiation of germ cells in XX mouse gonads occurs in an anterior-to-posterior wave. Dev. Biol., 262, 303–312. [DOI] [PubMed] [Google Scholar]
- 35. Wilhelm D., Washburn L.L., Truong V., Fellous M., Eicher E.M. and Koopman P. (2009) Antagonism of the testis- and ovary-determining pathways during ovotestis development in mice. Mech. Dev., 126, 324–336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Gonen N., Futtner C.R., Wood S., Garcia-Moreno S.A., Salamone I.M., Samson S.C., Sekido R., Poulat F., Maatouk D.M. and Lovell-Badge R. (2018) Sex reversal following deletion of a single distal enhancer of Sox9. Science (New York N.Y.), 360, 1469–1473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Schmahl J. and Capel B. (2003) Cell proliferation is necessary for the determination of male fate in the gonad. Dev. Biol., 258, 264–276. [DOI] [PubMed] [Google Scholar]
- 38. Schmahl J., Eicher E.M., Washburn L.L. and Capel B. (2000) Sry induces cell proliferation in the mouse gonad. Development, 127, 65–73. [DOI] [PubMed] [Google Scholar]
- 39. Nel-Themaat L., Vadakkan T.J., Wang Y., Dickinson M.E., Akiyama H. and Behringer R.R. (2009) Morphometric analysis of testis cord formation in Sox9-EGFP mice. Dev. Dyn., 238, 1100–1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Paralkar V.R., Taborda C.C., Huang P., Yao Y., Kossenkov A.V., Prasad R., Luan J., Davies J.O., Hughes J.R., Hardison R.C. et al. (2016) Unlinking an lncRNA from Its Associated cis Element. Mol. Cell, 62, 104–110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Krentz A.D., Murphy M.W., Zhang T., Sarver A.L., Jain S., Griswold M.D., Bardwell V.J. and Zarkower D. (2013) Interaction between DMRT1 function and genetic background modulates signaling and pluripotency to control tumor susceptibility in the fetal germ line. Dev. Biol., 377, 67–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Rahmoun M., Lavery R., Laurent-Chaballier S., Bellora N., Philip G.K., Rossitto M., Symon A., Pailhoux E., Cammas F., Chung J. et al. (2017) In mammalian foetal testes, SOX9 regulates expression of its target genes by binding to genomic regions with conserved signatures. Nucleic Acids Res., 45, 7191–7211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Minkina A., Matson C.K., Lindeman R.E., Ghyselinck N.B., Bardwell V.J. and Zarkower D. (2014) DMRT1 protects male gonadal cells from retinoid-dependent sexual transdifferentiation. Dev. Cell, 29, 511–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Buaas F.W., Val P. and Swain A. (2009) The transcription co-factor CITED2 functions during sex determination and early gonad development. Hum. Mol. Genet., 18, 2989–3001. [DOI] [PubMed] [Google Scholar]
- 45. Vidal V.P., Chaboissier M.C., Rooij D.G. and Schedl A. (2001) Sox9 induces testis development in XX transgenic mice. Nat. Genet., 28, 216–217. [DOI] [PubMed] [Google Scholar]
- 46. Chaboissier M.C., Kobayashi A., Vidal V.I., Lutzkendorf S., Kant H.J., Wegner M., de Rooij D.G., Behringer R.R. and Schedl A. (2004) Functional analysis of Sox8 and Sox9 during sex determination in the mouse. Development, 131, 1891–1901. [DOI] [PubMed] [Google Scholar]
- 47. Symon A. and Harley V. (2017) SOX9: A genomic view of tissue specific expression and action. Int. J. Biochem. Cell. Biol., 87, 18–22. [DOI] [PubMed] [Google Scholar]
- 48. Park M., Shin E., Won M., Kim J.H., Go H., Kim H.L., Ko J.J., Lee K. and Bae J. (2010) FOXL2 interacts with steroidogenic factor-1 (SF-1) and represses SF-1-induced CYP17 transcription in granulosa cells. Mol. Endocrinol., 24, 1024–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang D.-S., Kobayashi T., Zhou L.-Y., Paul-Prasanth B., Ijiri S., Sakai F., Okubo K., Morohashi K. and Nagahama Y. (2007) Foxl2 up-regulates aromatase gene transcription in a female-specific manner by binding to the promoter as well as interacting with Ad4 binding protein/steroidogenic factor 1. Mol. Endocrinol., 21, 712–725. [DOI] [PubMed] [Google Scholar]
- 50. Sekido R. and Lovell-Badge R. (2008) Sex determination involves synergistic action of SRY and SF1 on a specific Sox9 enhancer. Nature, 453, 930–934. [DOI] [PubMed] [Google Scholar]
- 51. Bullejos M. and Koopman P. (2001) Spatially dynamic expression of Sry in mouse genital ridges. Dev. Dyn., 221, 201–205. [DOI] [PubMed] [Google Scholar]
- 52. Katoh-Fukui Y., Miyabayashi K., Komatsu T., Owaki A., Baba T., Shima Y., Kidokoro T., Kanai Y., Schedl A., Wilhelm D. et al. (2012) Cbx2, a polycomb group gene, is required for Sry gene expression in mice. Endocrinology, 153, 913–924. [DOI] [PubMed] [Google Scholar]
- 53. Katoh-Fukui Y., Tsuchiya R., Shiroishi T., Nakahara Y., Hashimoto N., Noguchi K. and Higashinakagawa T. (1998) Male-to-female sex reversal in M33 mutant mice. Nature, 393, 688–692. [DOI] [PubMed] [Google Scholar]
- 54. Albrecht K.H. and Eicher E.M. (2001) Evidence that Sry is expressed in pre-Sertoli cells and Sertoli and granulosa cells have a common precursor. Dev. Biol., 240, 92–107. [DOI] [PubMed] [Google Scholar]
- 55. Mork L., Maatouk D.M., McMahon J.A., Guo J.J., Zhang P., McMahon A.P. and Capel B. (2012) Temporal differences in granulosa cell specification in the ovary reflect distinct follicle fates in mice. Biol. Reprod., 86, 37–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Pruitt S.C., Freeland A., Rusiniak M.E., Kunnev D. and Cady G.K. (2013) Cdkn1b overexpression in adult mice alters the balance between genome and tissue ageing. Nat. Commun., 4, 2626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Golson M.L. and Kaestner K.H. (2016) Fox transcription factors: from development to disease. Development, 143, 4558–4570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Benayoun B.A., Georges A.B., L'Hote D., Andersson N., Dipietromaria A., Todeschini A.L., Caburet S., Bazin C., Anttonen M. and Veitia R.A. (2011) Transcription factor FOXL2 protects granulosa cells from stress and delays cell cycle: role of its regulation by the SIRT1 deacetylase. Hum. Mol. Genet., 20, 1673–1686. [DOI] [PubMed] [Google Scholar]
- 59. Liu X.L., Meng Y.H., Wang J.L., Yang B.B., Zhang F. and Tang S.J. (2014) FOXL2 suppresses proliferation, invasion and promotes apoptosis of cervical cancer cells. Int. J. Clin. Exp. Pathol., 7, 1534–1543. [PMC free article] [PubMed] [Google Scholar]
- 60. Maatouk D.M., Mork L., Chassot A.A., Chaboissier M.C. and Capel B. (2013) Disruption of mitotic arrest precedes precocious differentiation and transdifferentiation of pregranulosa cells in the perinatal Wnt4 mutant ovary. Dev. Biol., 383, 295–306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Langmead B., Trapnell C., Pop M. and Salzberg S.L. (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol, 10, R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Heinz S., Benner C., Spann N., Bertolino E., Lin Y.C., Laslo P., Cheng J.X., Murre C., Singh H. and Glass C.K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol. Cell, 38, 576–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Sikora-Wohlfeld W., Ackermann M., Christodoulou E.G., Singaravelu K. and Beyer A. (2013) Assessing computational methods for transcription factor target gene identification based on ChIP-seq data. PLoS Comput. Biol., 9, e1003342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Madisen L., Zwingman T.A., Sunkin S.M., Oh S.W., Zariwala H.A., Gu H., Ng L.L., Palmiter R.D., Hawrylycz M.J., Jones A.R. et al. (2010) A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci., 13, 133–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


