Figure 1.
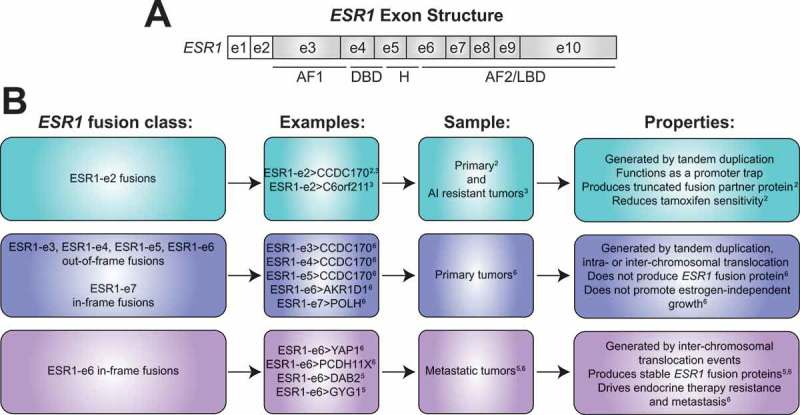
Landscape of ESR1 fusions in estrogen receptor positive breast cancer. (A) Structure of the estrogen receptor alpha gene (ESR1) containing 10 exons (e). The first two non shaded boxes represent non-coding exons (e1 and e2) followed by shaded boxes representing exons encoding for activation function 1 domain (AF1), DNA-binding domain (DBD), hinge region (H), and the activation function 2 (AF2)/ligand binding domain (LBD). (B) Three major classes of ESR1 fusion transcripts identified in estrogen receptor positive breast tumors. The first class arises from tandem duplication of ESR1promoter sequences including the first two exons of ESR1 (ESR1-e2) and a nearby gene found in primary and aromatase inhibitor (AI) resistant tumors, producing truncated forms of fusion partner protein, and therefore potentially driving disease pathogenesis by functioning as a promoter trap. The second class consists of the first 3–6 exons of ESR1 (ESR1-e3, ESR1-e4, ESR1-e5, ESR1-e6) fused out-of-frame or the first 7 exons of ESR1 (ESR1-e7) fused in-frame to C-terminal sequences from the partner gene. These fusion genes are found in treatment naïve primary breast tumors and do not produce stable ESR1 fusion proteins nor induce growth in estrogen-deprived conditions. The third class involves the first 6 exons of ESR1 (ESR1-e6) fused in-frame to C-terminal partner gene sequences provided by inter-chromosomal translocation found in metastatic breast tumors that have been extensively pretreated with endocrine agents. ESR1-e6>YAP1 and ESR1-e6>PCDH11X have been functionally characterized in detail and have been shown to drive endocrine therapy resistance and metastasis in experimental models. Coiled-coil domain containing protein 170, CCDC170; acidic residue methyltransferase 1, C6orf211; aldo-keto reductase family 1 member D1, AKR1D1; DNA polymerase eta, POLH; yes associated protein 1, YAP1; protocadherin 11 X-linked, PCDH11X; disabled homolog 2, DAB2; glycogenin 1, GYG1.
