Abstract
FOXP3, a lineage-specific forkhead (FKH) transcription factor, plays essential roles in the development and function of regulatory T cells. However, the DNA-binding properties of FOXP3 are not well understood. In this study, FOXP3 fragments containing different domains were purified, and their DNA-binding properties were investigated using electrophoretic mobility shift assay and isothermal titration calorimetry (ITC). Both the FKH and leucine-zipper domains were required for optimal DNA binding for FOXP3. FOXP3 protein not only binds with DNA sequences containing one FKH consensus sequence, but also binds with DNA sequences with two direct repeats of consensus sequences separated by three-nucleotides (DRE3). Our results shed lights on the mechanisms by which FOXP3 recognizes cognate DNA elements, and would facilitate further structural and functional studies of FOXP3.
Keywords: FOXP3, DNA binding, isothermal titration calorimetry, electrophoretic mobility shift assay, protein purification
Introduction
FOXP3, a forkhead winged-helix family (FOX) transcription factor, regulates the development and function of regulatory T cells (Treg) cells, which play critical roles in immune regulation and tolerance [1,2]. Deletion or mutation of FOXP3 causes autoimmunity in scurfy mice and IPEX syndrome (immune dysregulation, polyendocrinopathy, and enteropathy, X-linked) in human [3,4]. Ectopic FOXP3 expression induces a Treg phenotype in conventional CD4 + T cells, while targeted deletion of FOXP3 in CD4 + T cells of mice causes severe autoimmunity [5]. In addition, FOXP3 acts as a negative regulator of gene transcription by interaction with other transcription factors such as NFAT and AML1/Runx1 [6,7].
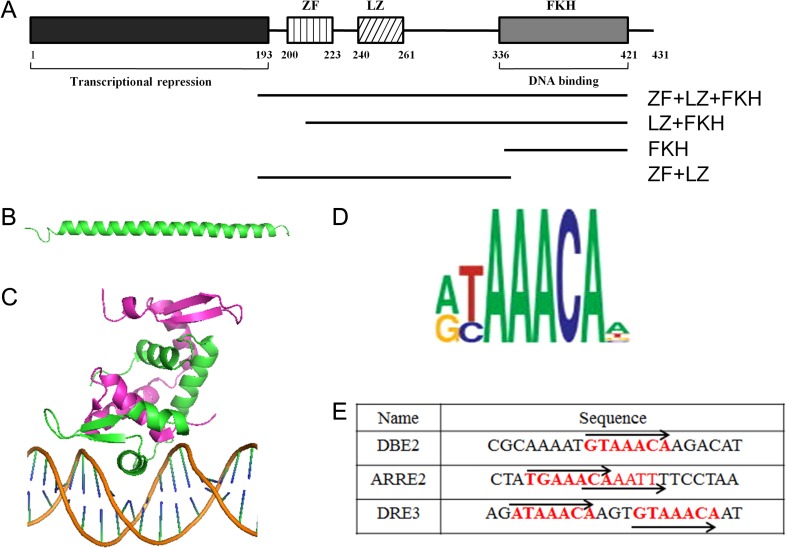
Human full-length FOXP3 protein contains an N-terminal domain required for transcriptional repression, a putative C2H2 zinc-finger (ZF) domain, a leucine-zipper (LZ) domain, and a C-terminal forkhead (FKH) domain (Fig. 1 A) [8]. The crystal structure of FOXP3 fragment containing the ZF and LZ regions (amino acids 196–276) has been solved [9]. The ZF loop (amino acids 196–209) is poorly defined, while the ZF helix and the LZ form an extended single long helix (Fig. 1B). The crystal structure of FOXP3-FKH domain has also been solved [10]. Two FOXP3-FKH domains form a domain-swapped dimer and bind with cognate DNA sequence (Fig. 1 C).
Figure 1.
FOXP3 domain structures and binding sequences (A) Domain composition of full-length FOXP3. (B) Structural representation of FOXP3-LZ + ZF fragment. The ZF loop was not defined in the structure, while the ZF helix and adjacent LZ domain formed an extended single long helix. (C) Structural representation of FOXP3-FKH domain in complex with DNA. Two FOXP3-FKH monomers form a domain-swapped dimer. (D) FOXP3 consensus-binding site. (E) Oligonucleotide sequence used in the EMSA experiments (the core recognition element ‘AAACA’ is shown in red). DBE2, Daf-16 family-binding element 2; ARRE2, a DNA sequence from human IL2 promoter; DRE3, a direct repeat of FKH consensus element separated by three-nucleotides.
FOXP3 regulates target gene expression by binding to its cognate sequences. Like most FKH transcription factors, FOXP3 has been shown to preferentially bind to the ‘GTAAACA’ motif (Fig. 1D) [11]. Koh et al. [12] showed that FOXP3 binds better with two FOXP3 consensus elements separated by three base-pairs, corresponding to one complete turn of the DNA helix. Since this site represents a direct repeat of two consensus elements separated by three-nucleotides, we named this site as DRE3 in this paper (Fig. 1E). FOXP3 can also associate with other transcription factors such as NFAT and AML1/Runx1 to regulate the transcription of specific subsets of FOXP3 target genes [13]. For example, FOXP3 and NFAT1 form a complex to bind to the ARRE2 sequence from the mouse IL-2 promoter [14].
Most FKH transcription factors use their FKH domain to bind with the FKH consensus site [15,16]. However, in our experience, the FKH domain of FOXP3 binds to the consensus site much weaklier than most other FKH transcription factors such as FOXO1 and FOXA2 (shown later), which suggested that other domain(s) of FOXP3 might be required for optimal DNA recognition. In this study, we successfully purified FOXP3 fragments containing different domains, and explored the domain requirements and sequence specificity of DNA binding for FOXP3.
Materials and Methods
Construction of expression vector
DNA fragments, encoding FKH domains of human FOXP3, FOXO1, and FOXA2, were cloned into pET28a vector (Invitrogen, Carlsbad, USA). Other FOXP3 fragments were inserted into pET28a or pMAL-c5x-p vectors, respectively. PMAL-c5x-p vector was modified from pMAL-c5x vector (Invitrogen), with the Factor Xa cleavage site substituted with PreScission cleavage site. All constructs were confirmed by DNA sequencing (GenScript, Nanjing, China).
Protein expression and purification
Proteins used in this study were all expressed in Escherichia coli. Proteins with His-tag were purified as described previously [17]. Proteins with maltose-binding protein (MBP)-tag were purified by amylose affinity chromatography. For the MBP-tagged proteins, the PreScission protease (inhouse produced) was used to remove the MBP-tag. Protein samples were further purified by Mono S cation exchange (GE Healthcare, Wisconsin, USA). The purity of each protein was analyzed by coomassie blue staining of SDS-PAGE (sodium dodecyl sulfate polyacrylamide gel electrophoresis) gel.
Electrophoretic mobility shift assay
Both protein and DNA samples were prepared at the concentration of 50 μM. DNA was incubated with protein for 30 min. A native 8% (w/v) polyacrylamide gel in 0.5× Tris borate EDTA (TBE) buffer was used to resolve the free DNA from the protein/DNA complex. The gel was visualized under ultraviolet light after being stained with ethidium bromide.
ITC experiments
Isothermal titration calorimetry (ITC) measurements were performed using NANO ITC (TA Instruments, New Castle, USA). DNA sample was dialyzed overnight against the storage buffer (25 mM HEPES, 250 mM NaCl, 10 mM MgCl2, and 1 mM TCEP, pH 7.0). Fifty microliter of DNA sample (100–200 μM) was injected to a FOXP3 protein solution (20 μM) in a 300-μl sample cell. The intervals between injections were 200 s, and the volume of injections at every turn was 2 μl. All data were analyzed using the software Launch NanoAnalyze (TA Instruments).
Bioinformatics
The DRE3 sequence RWAAAYANNNRWAAAYA (R = A/G, W = A/T, Y = C/T, N = any nucleotide) was searched using ChIP-seq peak sequences obtained from published ChIP-seq data [18].
Results
Protein expression and purification of FOXP3 fragments
FOXP3 contains multiple domains, including a putative ZF domain, a LZ domain, and a FKH domain (FKH), with LZ domain locating immediately adjacent to the ZF domain (Fig. 1 A). In order to determine the DNA-binding properties of FOXP3, we expressed multiple FOXP3 fragments containing different domains: FOXP3-FKH (amino acids 336–420), FOXP3-FKH + LZ (amino acids 205–420), FOXP3-ZF + LZ (amino acids 181–335), and FOXP3-FKH + LZ + ZF (amino acids 181–420) (Fig. 1 A). In the FKH + LZ fragment, the N-terminus was extended to the start position of the ZF helix, because previous studies showed that the ZF helix and the LZ domain form a long helix (Fig. 1B). By doing so, this fragment was able to keep the long helix intact, while the ZF function was lost because two of the four zinc-coordinated amino acids (Cys198 and Cys203) were absent in FOXP3-FKH + LZ fragment.
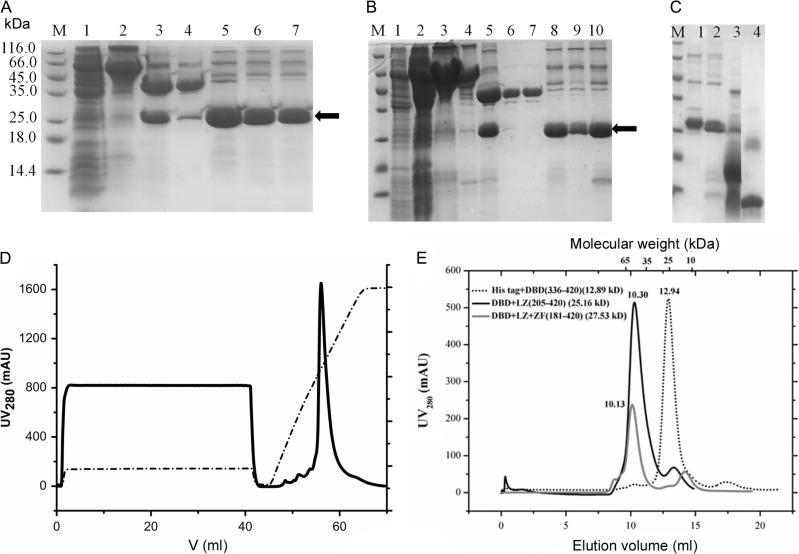
The FOXP3-FKH and FOXP3-LZ + ZF fragments were successfully expressed using His-tag and purified using Ni-NTA beads. The FOXP3-FKH + LZ and FOXP3-FKH + LZ + ZF fragments were insoluble with His-tag, but became soluble with MBP-tag. These two fragments were then expressed with MBP-tag, and purified using amylose affinity chromatography, followed by PreScission protease digestion to remove the MBP-tag (Fig. 2 A,B). To further improve the protein purity, all four FOXP3 fragments were subject to Mono S ion exchange chromatography. Purification of FOXP3-FKH + LZ using Mono S ion exchange chromatography was shown in Fig. 2D as an example. All four fragments were able to achieve 80%–90% purity after purification (Fig. 2 C).
Figure 2.
Expression and purification of different FOXP3 fragments (A) Purification of FOXP3-FKH + LZ + ZF fragment. Lane 1, supernatant of cell lysate; Lane 2, elution fractions from amylose-resin affinity column; Lane 3, after digestion with PreScission protease; Lane 4, flow-through fraction from mono S column; and Lanes 5–7, elution fraction from mono S column. (B) Purification of FOXP3-FKH + LZ fragment. Lane 1, supernatant of cell lysate; Lane 2, pellet of cell lysate; Lanes 3-4, elution fractions from amylose-resin affinity column; Lane 5, after digestion with PreScission protease; Lanes 6 and 7, flow-through fraction from mono S column; and Lanes 8–10, elution fractions from mono S column. (C) Purity analysis of different FOXP3 fragments. Lane 1, F FOXP3-FKH + LZ + ZF; Lane 2, FOXP3-FKH + LZ; Lane 3, FOXP3-LZ + ZF; and Lane 4, FOXP3-FKH. (D) Purification of FOXP3-FKH + LZ using Mono S 5/50 column. (E) Gel filtration elution profiles of different FOXP3 fragments using Superdex 75 10/300 column. The elution volumes of molecular size markers were reported on top of the figure. The elution volume of FOXP3 fragments suggested that all FOXP3 fragments are predominantly dimers in solution.
In order to study the oligomeric state of each fragment, size exclusion chromatography (Superdex 75 10/300 column) was employed. FOXP3-FKH, FOXP3-FKH + LZ, and FOXP3-FKH + LZ + ZF domains were shown to exist mainly in the dimer state in solution (Fig. 2E).
Domain requirement for optimized DNA binding
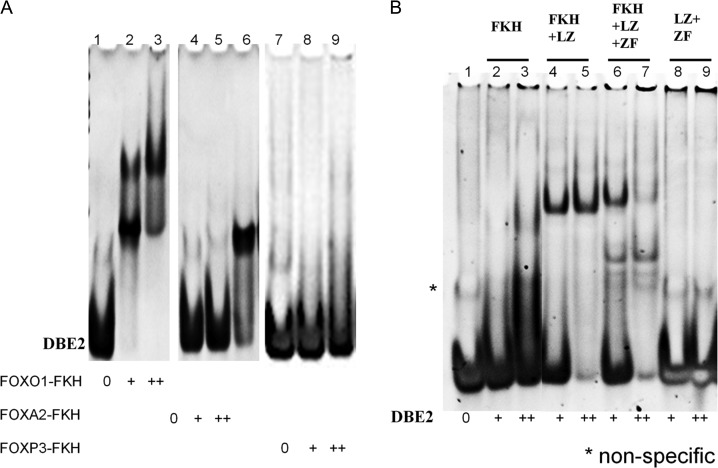
FKH transcription factors share a common DNA-binding FKH domain, which binds to a consensus DNA sequence (5′-GTAAACA-3′) [15,16]. Therefore, we compared the DNA-binding abilities of the FKH domains of three different FKH transcription factors (FOXO1, FOXA2, and FOXP3) to a known DNA sequence containing a single FKH consensus sequence (DBE2) (Fig. 3A). The FKH domain of FOXO1 had the strongest binding with a tendency to oligomerize. The FKH domain of FOXA2 showed moderate DNA binding as a monomer, while the FKH domain of FOXP3 only exhibited weak binding without a clear band (Fig. 3A). These results suggested that FOXP3 might need other domains for optimal DNA binding.
Figure 3.
DNA binding of FOXP3 with DBE2 DNA (A) Comparison of DBE2 DNA binding with FKH domains of FOXO1, FOXA2, and FOXP3. (B) Comparison of DBE2 DNA binding with different FOXP3 fragments.
To test this hypothesis, we compared the DNA binding of different FOXP3 fragments with DBE2 sequence. As shown in Fig. 3B, FOXP3-FKH and FOXP3-LZ + ZF showed little or no binding to DBE2 sequence (Lanes 2–3 and 8–9). Both FOXP3-FKH + LZ + ZF and FOXP3-FKH + LZ bind to DBE2 DNA more stronger than FOXP3-FKH and FOXP3-LZ + ZF (Fig. 3B). Two bands of protein/DNA complexes were observed with FOXP3-FKH + LZ + ZF fragment, while only one band of protein/DNA complex were observed with FOXP3-FKH + LZ fragment. Based on the shifting patterns, we speculated that FOXP3-FKH + LZ + ZF fragment could bind with DBE2 DNA as both monomer and dimer, while FOXP3-FKH + LZ fragment could bind with DBE2 DNA mainly as dimer.
Quantitative analysis of DNA binding
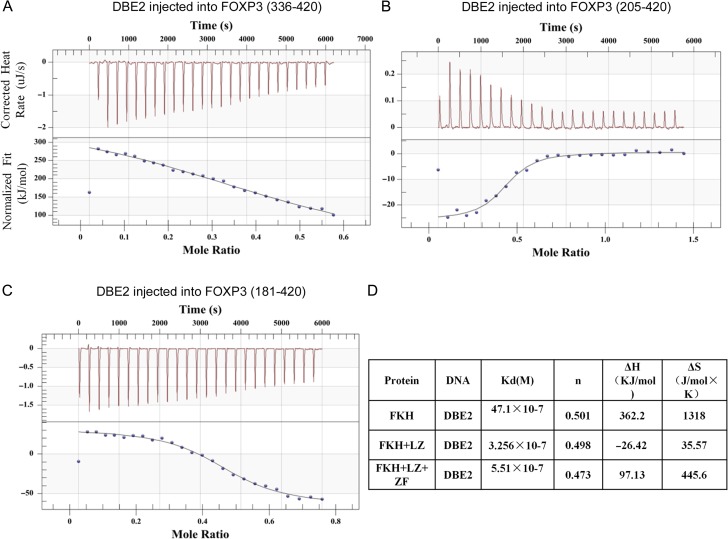
Quantitative analysis of DNA binding with different FOXP3 fragments was carried out using ITC. DBE2 DNA was injected into various FOXP3 fragments, and the heat generated or consumed by the binding reaction was measured (Fig. 4A–C). Upon DBE2 binding, different FOXP3 fragments exhibited the same stoichiometric ratio (n ≈ 0.5). FOXP3-FKH fragment bound to DBE2 DNA weakly with a Kd of 4.71 μM. FOXP3-FKH + LZ and FOXP3-FKH + LZ + ZF fragments bound to DBE2 DNA with much higher affinity with Kd of 0.33 and 0.55 μM, respectively (Fig. 4D). These results, together with electrophoretic mobility shift assay (EMSA) results, strongly suggested that both LZ and FKH domains of FOXP3 were required for optimal DNA binding, while ZF domain seemed to be dispensable for the binding of FOXP3 with DNA.
Figure 4.
Quantitative analysis of DBE2 DNA with different FOXP3 domain using ITC (A–C) Representative ITC isotherms of the binding of DBE2 DNA with different FOXP3 fragments. The raw curves showed the change in thermal power with regard to time in the period of titration (top). The bottom curves showed the heat of reaction normalized with the molar ratio. Standard free energies of binding and entropic contributions were also obtained from the bottom curves. (D) Thermodynamic parameters of the interaction between FOXP3 fragments and DBE2 DNA. All data were measured at 298 K in 25 mM HEPES, pH 7.5, 250 mM NaCl, 10 mM MgCl2, and 1 mM TCEP.
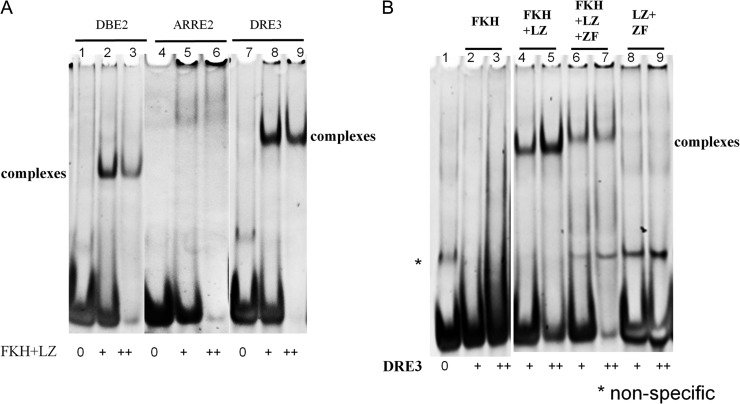
Binding of FOXP3 with different DNA sequences
We have demonstrated that both FKH and LZ domains are crucial for binding of FOXP3 with DNA. We then used FOXP3-FKH + LZ fragment to test its binding with three different DNA sequences: a DBE2 sequence containing a FKH consensus element, an ARRE2 sequence which has been reported to be bound by a FOXP3 domain-swapped dimer, and a DRE3 sequence containing a direct repeat of two consensus elements separated by three-nucleotides (Fig. 1E). Based on the decrease of free DNA upon protein binding, FOXP3-FKH + LZ fragment bound to all three DNA sequences, but with different-binding patterns (Fig. 5A). Upon binding to DRE3 sequence, FOXP3-FKH + LZ fragment seemed to form a higher order of oligomer than FOXP3-FKH + LZ/DBE2 complex, presumably as a tetramer (Fig. 5A, Lanes 8–9). When bound to the ARRE2 sequence, FOXP3-FKH + LZ fragment seemed to form an even higher order of oligomers than with the DRE3 sequence, without a major band in the EMSA gel (Fig. 5A, Lanes 5–6).
Figure 5.
DNA binding of FOXP3 with DRE3 DNA (A) FOXP3-FKH + LZ fragment bound to different DNA sequences: DBE2, ARRE2, and DRE3. (B) DNA binding of DRE3 DNA with different FOXP3 fragments.
Then, the binding of DRE3 sequence was assessed using different FOXP3 fragments. FOXP3-FKH and FOXP3-LZ + ZF fragments showed little or no binding with DRE3 DNA (Fig. 5B, Lanes 2–3 and 8–9). FOXP3-FKH + LZ fragment bound to DRE3 sequence in a similar pattern as FOXP3-FKH + LZ + ZF fragment (Fig. 5B, Lanes 4–5 and 6–7). These results were consistent with the findings in DBE2 DNA.
Identification of DRE3 sites in human genome
Previous ChIP-seq studies have revealed the FOXP3-binding sites in genome, and identified that FOXP3 binds to a consensus motif (RWAAAYA, R = A/G; W = T/A; and Y = C/T) in genome [18–20]. Our studies and other studies have demonstrated that FOXP3 could bind to the DRE3 sequence which contained two consensus FKH motifs. To the best of our knowledge, the existence of such FOXP3-binding DRE3 sequence in the genome has not been reported. Therefore, we analyzed the Samstein's ChIP-seq data and looked for the existence of DRE3-like sequences among FOXP3-binding sequences [18]. Several putative DRE3 sites were identified at the promoters of STAT6, IGF1R, CD79b, and CD22, and at the introns of FOXP1, ETS1, and IKZF2 (Table 1). Many of these genes play important roles in the function of Treg cells [21], indicating that the DRE3-like sites might be physiologically important for FOXP3’s function in regulating Treg cells.
Table 1.
Putative FOXP3-binding DRE3 sequences in human genome
 |
|---|
 |
The imperfect position is shown in lower-case letter, and the core binding sites of DRE3 sequence are colored in red.
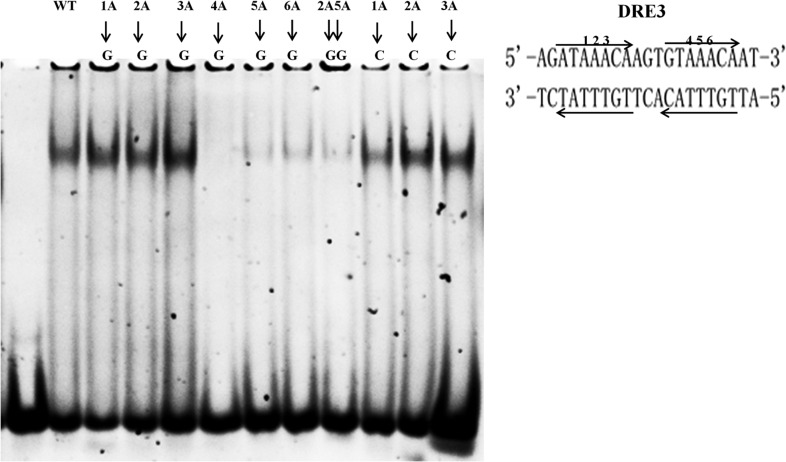
Mutation studies of DRE3 sequence
Previous studies have shown that the middle-three adenines within the ‘RWAAAYA’ (underlined) are important for FOXP3 recognition. In order to further investigate the DNA-binding properties of FOXP3 protein with DRE3 sequence, we tested the binding of FOXP3-FKH + LZ fragment with different DNA variants with substitutions of the middle-three adenines (Fig. 6). Surprisingly, our results showed that the second consensus site of the DRE3 sequence seemed to be more important for FOXP3 DNA recognition. When one of the three adenines was mutated within the first consensus site, FOXP3-FKH + LZ fragment could still bind with the DNA variant. However, when one of the three adenines was mutated within the second consensus site, the DNA binding was greatly diminished (Fig. 6). To our surprise, when the second consensus site was mutated, FOXP3-FKH + LZ fragment did not form a lower order of oligomer with DNA, even though the DNA variant kept an intact consensus site (Fig. 6). These results suggested that FOXP3 might recognize DRE3-like sequences with an imperfect consensus site followed by a perfect consensus site.
Figure 6.
DNA binding of DRE3 DNA variants with FOXP3-FKH + LZ fragment Nucleotide substitution was indicated using arrows.
Discussion
FOXP3 regulates the transcriptional profile of Treg cells, and inhibits spontaneous autoimmunity. In order to study the DNA-binding properties of FOXP3, we tried to express multiple FOXP3 fragments. Small FOXP3 fragments (FOXP3-FKH and FOXP3-LZ + ZF) were successfully expressed as His-tag fusion proteins. However, large fragments of FOXP3 (FOXP3-FKH + LZ + ZF and FOXP3-FKH + LZ) were insoluble with His-tag. We then tried to fuse these FOXP3 large fragments with MBP-tag, and found that MBP-tag could remarkably enhance the solubility of these FOXP3 large fragments. Compared with His-tag, MBP-tag has been reported to enhance the solubility of its fusion partner [22–24].
Previous ChIP-seq studies have revealed that FOXP3 can bind to sequences containing a FKH consensus sequence (5′-GTAAACA-3′) [11]. In this study, FOXP3-FKH domain was shown to have much weaker binding to DBE2 sequence than the FKH domains of FOXO1 and FOXA2. The weaker binding ability could be attributed to two unique features of FOXP3-FKH domain. First, FOXP3-FKH domain forms stable domain-swapped dimer in solution. Second, FOXP3-FKH domain has a shorter wing region than most other FKH transcription factors [10]. Therefore, FOXP3 might need more than its FKH domain to bind with its cognate DNA. FOXP3-FKH + LZ fragment, which contains both LZ and FKH domains, showed much better DNA binding than FKH domain alone. Further inclusion of the ZF domain did not increase the DNA binding. Although DBE2 only contains one FKH consensus sequence, FOXP3-FKH + LZ fragment seems to bind with DBE2 as a dimer. This is not surprising because of two reasons. First, the FKH domain of FOXP3 has been shown to form a domain-swapped dimer [10]. Second, the FOXP3-LZ domain has been shown to oligomerize [9].
In addition to the single FKH consensus sequence, FOXP3 could bind with sequences containing two FKH consensus sites. Koh et al. [12] showed that FOXP3 could bind with two consensus sites on the same side of the DNA. Using high-throughput SELEX sequencing, Jolma et al. [25] showed that FOXP3 could bind with a direct repeat of two consensus FKH sites separated DRE3. In this study, we showed that FOXP3 could efficiently bind with DRE3 site. Based on the shift pattern, we speculated that FOXP3 could bind with DRE3 site as a tetramer. Interestingly, Greene's group found that FOXP3 indeed could form a tetramer [26]. Another example of transcription factor tetramer is tumor suppressor p53 [27]. Similar to FOXP3, p53 also has a DNA-binding domain and an oligomerization domain. Unlike FOXP3, the oligomerization domain of p53 locates at the C-terminal of the DNA-binding domain. In addition, the linker region between the two domains in p53 is much shorter than that in FOXP3.
In order to test whether DRE3 sites are indeed bound by FOXP3 protein in the cell, we searched for DRE3 sites in the previously published ChIP-seq data. Putative DRE3 sites were found within the promoters of STAT6, IGF1R, CD79b, and CD22, and within the introns of FOXP1, ETS1, and IKZF2. Many of these gene products play key roles in regulating immune responses. Based on the results of the binding of FOXP3-FKH + LZ fragment with the DRE3 DNA variants, we found that the second consensus site within the DRE3 sequence was more important than the first one.
In summary, both the FKH and LZ domains are required for the optimal DNA binding for FOXP3. FOXP3 not only binds with DNA sequences containing a single FKH consensus site, but also binds with DNA sequences containing two FKH consensus sites separated by three-nucleotides.
Funding
This work was supported by the grants from National Natural Science Foundation of China (Nos. 81372904, 81570537 to Y.C. and No. 81272971 to Z.C.).
References
- 1. Zheng Y, Rudensky A. Foxp3 in control of the regulatory T cell lineage. Nat Immunol 2007, 8: 457–462. [DOI] [PubMed] [Google Scholar]
- 2. Park J, Ko JS, Shin Y, Cho JY, Oh H, Bothwell AM, Lee S. Intranuclear interactomic inhibition of FoxP3 suppresses functions of Treg cells. Biochem Biophys Res Commun 2014, 451: 1–7. [DOI] [PubMed] [Google Scholar]
- 3. Zhang W, Sharma R, Ju S, He X, Tao Y, Tsuneyama K, Tian Z, et al. Deficiency in regulatory T cells results in development of antimitochondrial antibodies and autoimmune cholangitis. Hepatology 2009, 49: 545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Joly A, Liu S, Dahlberg CIM, Mailer RKW, Westerberg LS, Andersson J. Foxp3 lacking exons 2 and 7 is unable to confer suppressive ability to regulatory T cells in vivo. J Autoimmun 2015, 63: 23–30. [DOI] [PubMed] [Google Scholar]
- 5. Buckner JH, Ziegler SF. Functional analysis of FOXP3. Ann N Y Acad Sci 2008, 1143: 151–169. [DOI] [PubMed] [Google Scholar]
- 6. Lopes JE, Torgerson TR, Schubert LA, Anover SD, Ocheltree EL, Ochs HD, Ziegler SF. Analysis of FOXP3 reveals multiple domains required for its function as a transcriptional repressor. J Immunol 2006, 5: 3133–3142. [DOI] [PubMed] [Google Scholar]
- 7. Marson A, Kretschmer K, Frampton GM, Jacobsen ES, Polansky JK, MacIsaac KD, Levine SS, et al. Foxp3 occupancy and regulation of key target genes during T-cell stimulation. Nature 2007, 445: 931–935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Lozano T, Casares N, Lasarte JJ. Searching for the achilles heel of FOXP3. Front Oncol 2013, 3: 00294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Song X, Li B, Xiao Y, Chen C, Wang Q, Liu Y, Berezov A, et al. Structural and biological features of FOXP3 dimerization relevant to regulatory T cell function. Cell Rep 2012, 1: 665–675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chen Y, Chen C, Zhang Z, Liu CC, Johnson ME, Espinoza CA, Edsall LE, et al. DNA binding by FOXP3 domain-swapped dimer suggests mechanisms of long-range chromosomal interactions. Nucleic Acids Res 2015, 43: 1268–1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sadlon TJ, Wilkinson BG, Pederson S, Brown CY, Bresatz S, Gargett T, Melville EL. Genome-wide identification of human FOXP3 target genes in natural regulatory T cells. J Immunol 2010, 2: 1071–1081. [DOI] [PubMed] [Google Scholar]
- 12. Koh KP, Sundrud MS, Rao A. Domain requirements and sequence specificity of DNA binding for the forkhead transcription factor FOXP3. PLos One 2009, 12: e8109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Ono M, Yaguchi H, Ohkura N, Kitabayashi I, Nagamura Y, Nomura T, Miyachi Y, et al. Foxp3 controls regulatory T-cell function by interacting with AML1/Runx1. Nuature 2007, 446: 685–689. [DOI] [PubMed] [Google Scholar]
- 14. Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, et al. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell 2006, 126: 375–387. [DOI] [PubMed] [Google Scholar]
- 15. Brent MM, Anand R, Marmorstein R. Structural basis for DNA recognition by FoxO1 and its regulation by posttranslational modification. Structure 2008, 16: 1407–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Littler DR, Alvarez-Fernandez M, Stein A, Hibbert RG, Heidebrecht T, Aloy P, Medema RH, et al. Structure of the FoxM1 DNA-recognition domain bound to a promoter sequence. Nucleic Acids Res 2010, 38: 4527–4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wu D, Guo M, Philips MA, Qu L, Jiang L, Li J, Chen X, et al. Crystal structure of the FGFR4/LY2874455 complex reveals insights into the Pan-FGFR selectivity of LY2874455. PLoS One 2016, 11: e0162491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Samstein R, Arvey A, Josefowicz S, Peng X, Reynolds A. Foxp3 exploits a pre-existent enhancer landscape for regulatory T cell lineage specification. Cell 2012, 151: 153–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang B, Lin D, Li C, Tucker P. Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. J Biol Chem 2003, 278: 24259–24268. [DOI] [PubMed] [Google Scholar]
- 20. Nelson CS, Fuller CK, Fordyce PM, Greninger AL, Li H, DeRisi JL. Microfluidic affinity and ChIP-seq analyses converge on a conserved FOXP2-binding motif in chimp and human, which enables the detection of evolutionarily novel targets. Nucleic Acids Res 2013, 41: 5991–6004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Rudra D, DeRoos P, Chaudhry A, Niec RE, Arvey A, Samstein RM, Leslie C, et al. Transcription factor Foxp3 and its protein partners form a complex regulatory network. Nat Immunol 2012, 13: 1010–1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Xiao H, Shi Y, Yuan J, Huang Y, Wang J. Over-expression, rapid preparation and some properties of C-terminal BARc region in PICK1. Int J Mol Sci 2009, 10: 28–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hewitt SN, Choi R, Kelley A, Crowther GJ, Napuli AJ, Van Voorhis WC. Expression of proteins in Escherichia coli as fusions with maltose-binding protein to rescue non-expressed targets in a high-throughput protein-expression and purification pipeline. Acta Crystallogr Sect F Struct Biol Cryst Commun 2011, 67: 1006–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Raran-Kurussi S, Waugh DS. The ability to enhance the solubility of its fusion partners is an intrinsic property of maltose-binding protein but their folding is either spontaneous or chaperone-mediated. PLoS One 2012, 7: e49589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jolma A, Yan J, Whitington T, Toivonen J, Nitta KR, Rastas P, Morgunova E, et al. DNA-binding specificities of human transcription factors. Cell 2013, 152: 327–339. [DOI] [PubMed] [Google Scholar]
- 26. Fontenot JD, Rasmussen JP, Gavin MA, Rudensky AY. A function for interleukin 2 in Foxp3-expressing regulatory T cells. Nat Immunol 2005, 6: 1142–1151. [DOI] [PubMed] [Google Scholar]
- 27. Chen Y, Dey R, Chen L. Crystal structure of the p53 core domain bound to a full consensus site as a self-assembled tetramer. Structure 2010, 18: 246–256. [DOI] [PMC free article] [PubMed] [Google Scholar]