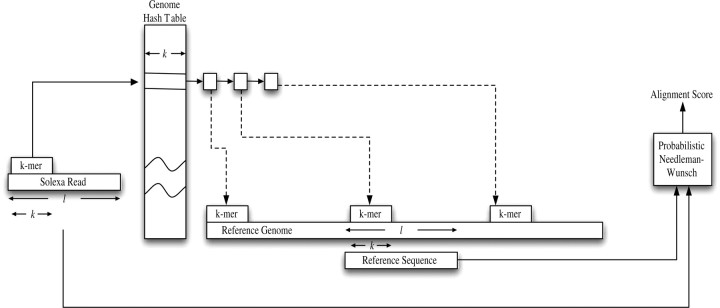
Fig. 1.
A flow-chart of the GNUMAP algorithm. First, the algorithm will incrementally find a k-mer piece in the consensus Solexa read. This k-mer is used as an index into the hash table, producing a list of positions in the genome with the exact k-mer sequence. These locations are expanded to align the same l nucleotides from the read to the genomic location. If the alignment score passes the user-defined threshold, the location is considered a hit, and recorded on the genome for future output.