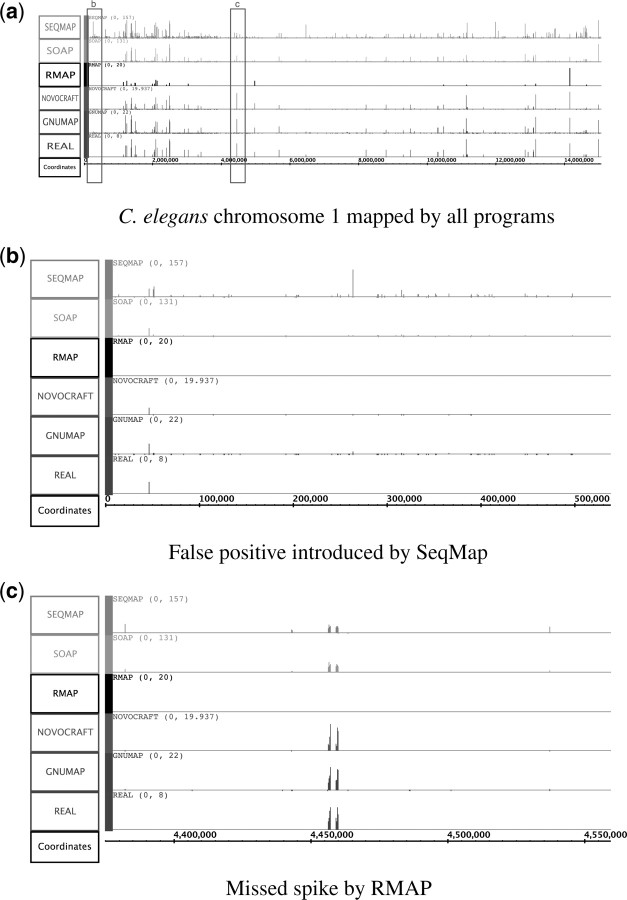
Fig. 4.
Benchmark real spikes (bottom) compared with SeqMap (Jiang and Wong, 2008), SOAP (Li,R. et al., 2008), RMAP (Smith et al., 2008), Novocraft (unpublished data) and GNUMAP. Benchmark data were constructed by sampling from 1000 promoter regions in the C.elegans genome. In (b) [an enlargement of the first boxed region in (a)], SeqMap incremented every location for reads from identical regions, producing a significant false positive spike. Attempting to remove false positives by discarding these reads, such as was done by RMAP, results in missing important information, as can be seen in (c) [an enlargement of the second boxed region in (a)]. Note: The intention of this figure is not to discuss the relative mapping capabilities of all currently implemented programs specifically, but to show the trend that would occur if each of these read-placement techniques were used.