Abstract
Previously, we identified SETD2 loss-of-function mutations in 22% of MLL-rearranged (MLLr) acute leukemia patients, implicating a mechanism for cooperativity between SETD2 mutations and MLL fusions. However, the detailed mechanism of how SETD2-H3K36me3 downregulation accelerates MLLr leukemia remains unclear. Here, we show that in MLLr leukemia, both H3K79me2 and H3K36me3 are aberrantly elevated and co-enriched in a group of genes. SETD2 inactivation leads to a global reduction of H3K36me3 and a further elevation of H3K79me2, but does not change the expression of known MLL fusiontarget genes. Instead, this pattern of histone changes is associated with transcriptional deregulation of a novel set of genes; downregulating tumor suppressors (for example, ASXL1) and upregulating oncogenes (for example, ERG). Taken together, our findings reveal a global crosstalk between the oncogenic DOT1L-H3K79me2 axis and the tumor suppressive SETD2-H3K36me3 axis in gene regulation, provide molecular insights into how SETD2 mutations accelerate MLLr leukemogenesis through differential regulation of additional tumor suppressors and oncogenes.
INTRODUCTION
The SET domain containing 2 (SETD2) gene encodes the histone 3 lysine 36 (H3K36) methyltransferase which is responsible for trimethylation (H3K36me3).1 SETD2 loss-of-function (LOF) mutations have been identified in various tumor types, indicating the tumor suppressor function of SETD2.2–4 We previously identified about 6% of somatic SETD2 mutations in patients with acute myeloid leukemia (AML) and acute lymphoid leukemia (ALL). Moreover, SETD2 mutations were enriched in about 22% of MLL-rearranged (MLLr) leukemia patients.5 Two additional reports have also shown a similar mutation frequency of SETD2 in MLLr patients.6,7 Using an Mll-AF9 knock-in (MA9) AML mouse model, we found that knockdown (KD) of Setd2 could significantly accelerate disease development.5 All of these findings indicate a strong cooperation between SETD2 inactivation and MLL fusions; however, the detailed mechanism remains unknown.
SETD2 is the main methyltransferase generating H3K36me3.1 Thus SETD2 LOF could reduce genome-wide H3K36me3. The high mutation frequency of SETD2 in MLLr leukemia also implicates the cooperation of SETD2-mediated H3K36me3 changes and MLL fusions. H3K36me3 is a histone mark enriched at gene bodies of transcriptionally active genes and modified along with transcription elongation of phosphorylated RNA polymerase II.8–11Genome-wide H3K36me3 change could induce large-scale gene dysregulation, which may be related to leukemogenesis. However this epigenetic mechanism is poorly understood. Uncovering this mechanism would lead to a better understanding of why SETD2 is selected for mutation, and how SETD2-H3K36me3 downregulation contributes to leukemia development.
Another well-studied elongation-related mark is histone H3 lysine 79 dimethylation (H3K79me2), which is also enriched at gene bodies and correlated with transcription activation.11–13 In MLLr leukemia, MLL fusion proteins could recruit H3K79 methyltransferase DOT1L to MLL fusion gene targets, and induce an aberrantly high level of H3K79me2 which results in gene activation and leukemogenesis. High expression levels of MLL fusion gene targets, such as Hoxa9, Mecom and Meis1, have been well documented to be driven by aberrant H3K79me2.14–16 However, it is still unclear whether this single histone change is sufficient to promote leukemogenesis and how it may affect the landscape of the epigenome in pre-leukemia and in leukemia. As two frequently dysregulated elongation marks in leukemia patients, there may be potential crosstalk between H3K79me2 and H3K36me3. We aim to understand whether gene body H3K79me2 changes will affect H3K36me3 and whether this potential H3K36me3 change contributes to gene dysregulation in MLLr leukemia without SETD2 LOF. Furthermore, whether downregulation of SETD2-H3K36me3 axis could affect DOT1LH3K79me2 axis in MLLr leukemia with SETD2 LOF, and whether this potential crosstalk could contribute to MLLr leukemia progression.
In this study, we explored the epigenetic crosstalk between H3K79me2 and H3K36me3, and gene regulation modulated by histone changes in MLLr leukemia under SETD2 wild-type (SETD2-WT) and SETD2 LOF conditions. We found a dynamic gene regulation crosstalk between oncogenic DOT1L-H3K79me2 and tumor suppressive SETD2-H3K36me3 modulations which promotes leukemogenesis through differential regulation of additional subsets of tumor suppressor genes (TSGs) and oncogenes. We further validated that both downregulation of TSG ASXL1 and upregulation of oncogene ERG contribute to SETD2-H3K36me3 loss-mediated leukemia acceleration.
MATERIALS AND METHODS
Animals
The Asxl1fl/fl mice were purchased from Jackson Laboratories, and bred with Mx1-Cre mice. To generate Asxl1Δ/Δ mice, the Mx1-Cre-Asxl1fl/fl conditional mice received five intraperitoneal injections of polyI:polyC every other day at a dose of 10 mg/kg of body weight.
Immuno-blotting
Samples used for immuno-blotting were extracted using home-made 1 × SDS sample buffer. Signal quantification was performed using ImageJ software. For each antibody, we did multiple repeat experiments and only representative results are shown.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed using home-made reagents. ChIP DNA libraries were conducted with ChIP-seq kit (Bioo Scientific, 5143–02) according to the protocol. Then the quality control (QC) passed libraries were subjected to sequencing using Hiseq2000 platform.
Detailed materials and methods are described in supplementary information.
RESULTS
Global increase of H3K36me3 by MLL-AF9
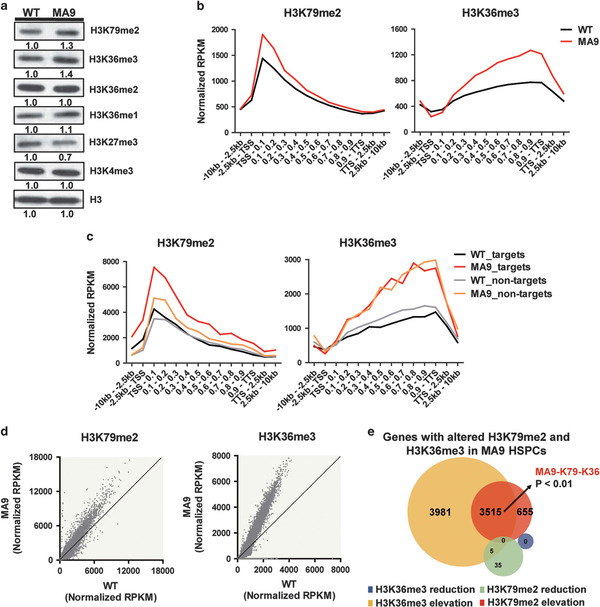
H3K79me2 and H3K36me3 are two well-studied transcriptional elongation-related histone marks enriched at active gene bodies (Supplementary Figure S1a). To explore the potential crosstalk between these two marks, we first investigated whether the increased H3K79me2 in MLLr leukemia could affect the H3K36me3 levels when there is no genetic alteration for SETD2. Using immuno-blotting, we measured the global levels of these two histone marks in c-Kit-positive (c-Kit+) hematopoiesis stem/progenitor cells (HSPCs) isolated from MA9 and C57/BL/6 wild-type (WT) mice. Interestingly, both H3K79me2 and H3K36me3 were increased in MA9 HSPCs compared to WT HSPCs (Figure 1a). To further investigate the genome-wide changes of these two marks, we performed ChIP-seq of H3K79me2 and H3K36me3 in WT and MA9 HSPCs. H3K79me2 and H3K36me3 were preferentially enriched at gene bodies, but no significant changes of peak distributions were found (Supplementary Figure S1b). Consistent with published studies,15 a significant global increase of H3K79me2 at gene bodies was found in MA9 HSPCs compared to WT HSPCs. Surprisingly, a significant increase of H3K36me3 at gene bodies was also observed (Figure 1b, Supplementary Figure S1c). This suggests that there is an enhanced activity of tumor suppressor SETD2 in MA9 cells with increased oncogenic DOT1L-H3K79me2 axis.
Figure 1.
Increased H3K79me2 and H3K36me3 levels in MA9 HSPCs. (a) Histone changes detected by immuno-blotting. (b) ChIP-seq profiles of H3K79me2 and H3K36me3 for gene bodies (left: **P<0.01; right: ***P<0.001) in WT and MA9 HSPCs. P-values were calculated by two-tailed Student’s t test. (c) H3K79me2 and H3K36me3 profiles of selected MLL-AF9 targets (targets) (left: ***P<0.001; right: **P<0.01) and non-targets (left: ***P<0.001; right ***P<0.001) for gene bodies in WT and MA9 HSPCs. (d) H3K79me2 and H3K36me3 ChIP signals on each gene in MA9 compared to WT. (e) Venn diagram of genes with altered H3K79me2 and/or H3K36me3 occupancies at gene bodies. The P-value of the overlapped genes was calculated by hypergeometric test (**P<0.01). See also Supplementary Table S1.
Considering the aberrant H3K79me2 increase was known to be most prominent on MLL fusion target genes,15 we further studied whether H3K36me3 would show the same preferential enrichment on MLL-AF9 targets or non-MLL-AF9 targets (non-targets) (MLL-AF9 targets and non-targets were previously defined in published papers, the non-targets were randomly picked genes at similar expression levels with MLL-AF9 targets15,17). Although both H3K79me2 and H3K36me3 were increased, they showed different patterns. The increase of H3K79me2 was much more significant on MLL-AF9 targets. However, the increase of H3K36me3 was global and not specific on MLL-AF9 targets (Figure 1c). We found consistent results using published ChIP-seq data15 (Supplementary Figure S1d). Similar patterns were detected using another MLL-ENL targets set we identified in our previous work14 (Supplementary Figure S1e). These results indicate that the increase of H3K79me2 and H3K36me3 are not limited to MLLAF9 but may also be applied to other MLL fusions.
To explore the potential target genes modulated by increased H3K79me2 and H3K36me3, we determined the genes with altered changes of these two marks. Almost all of the genes (8151/8191) with histone modification changes showed increased H3K79me2 and/or H3K36me3 in MA9 compared to WT HSPCs. We defined these genes as H3K79me2 and H3K36me3 targets in MA9 HSPCs respectively (Figures 1d and e, Supplementary Table S1). The H3K79me2 targets showed a high level of consistency with the published data set (Supplementary Figure S2a). Interestingly, we found that H3K79me2 targets significantly overlapped with H3K36me3 targets, suggesting these two marks were co-enriched in, and may also co-regulate, a large set of genes. These overlapped genes were identified as H3K79me2 and H3K36me3 co-enriched genes in MA9 HSPCs (MA9-K79-K36 genes) (Figure 1e, Supplementary Table S1).
Gene dysregulation in MA9 HSPCs with increased H3K36me3 besides increased H3K79me2
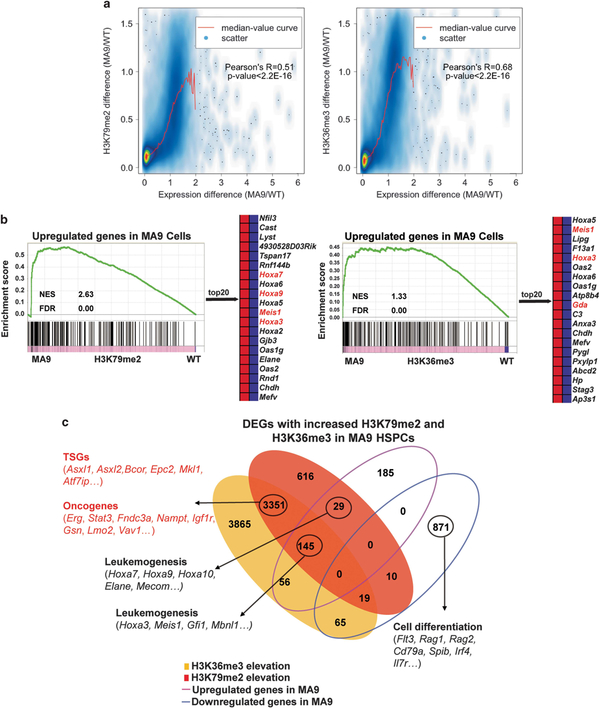
To explore whether increased H3K79me2 and H3K36me3 have an impact on gene expression, we performed RNA-seq using HSPCs from MA9 and WT mice. Through integrated ChIP-seq and RNA-seq analysis, we found that changes in H3K79me2 and H3K36me3 were positively correlated with differences in gene expression respectively (Figure 2a, Supplementary Figure S2b). Furthermore, gene set enrichment analysis (GSEA) revealed that upregulated genes in MA9 HSPCs had a coordinated increase in both H3K79me2 and H3K36me3 (Figure 2b), supporting that gene dysregulation in MA9 HSPCs is related to the increase of both H3K79me2 and H3K36me3. Only less than 10% (94/965) of the downregulated genes showed histone changes. By contrast, over 55% (230/415) of the upregulated genes in MA9 HSPCs showed an increase of H3K79me2 or H3K36me3 (Figure 2c). The downregulated genes may be related to differentiation blockage; the upregulated genes include MLL-AF9 targets Hoxa9 and Meis1. Interestingly, the largest group of MA9-K79-K36 genes (3351 genes) showed no expression change. Among them, there were many TSGs and oncogenes which have been reported in leukemogenesis (Figures 2b and c, Supplementary Figure S2c, Supplementary Table S2). We speculate that this group of genes may have been epigenetically marked by H3K79me2 and H3K36me3 in MLLr leukemia, and are susceptible to changes in mRNA expression upon further alteration on the SETD2-H3K36me3 axis.
Figure 2.
Gene dysregulation in MA9 HSPCs with increased H3K79me2 and H3K36me3 levels. (a) The correlation between H3K79me2 and H3K36me3 differences and expression changes in MA9 compared to WT HSPCs. Pearson’s correlation and two-tailed Student’s t-test (P-value) were calculated. (b) GSEA showing enrichment of upregulated genes at genome-wide gene body H3K79me2 and H3K36me3 ChIP signals respectively. MLL-AF9 targets are shown in red, non-targets are shown in black (NES: normalized enrichment score). (c) Venn diagram of differentially expressed genes (DEGs) with altered H3K79me2 or/and H3K36me3 occupancies. Genes in red show genes with no significant expression change, but were co-enriched with increased H3K79me2 and H3K36me3. See also Supplementary Tables S1 and S2.
SETD2-H3K36me3 loss-of-function further elevated H3K79me2
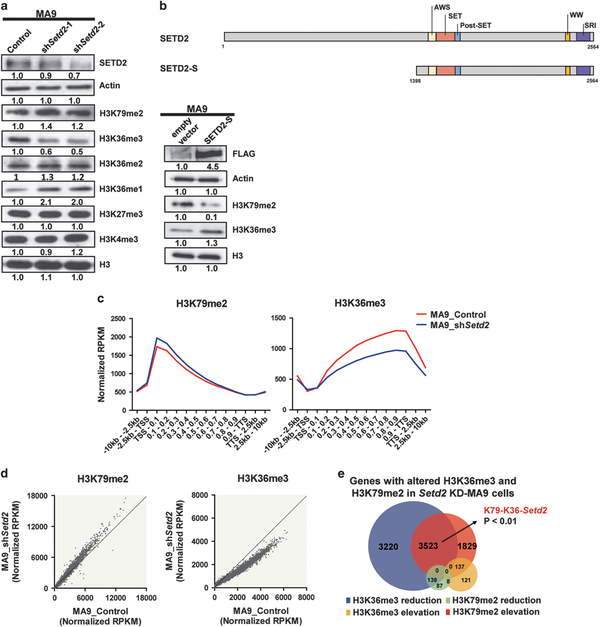
To explore whether the decreased H3K36me3 mediated by SETD2 LOF could further affect H3K79me2 levels and transcriptional activities in MLLr leukemia, we first mimicked the SETD2 LOF with MLL fusions in patients by performing KD of Setd2 in MA9 HSPCs. Enhanced proliferation and self-renewal capacities were observed (Supplementary Figure S3a). Interestingly, an increased H3K79me2 level was also observed besides a dramatic decrease of H3K36me3 in MA9 cells after Setd2 KD (Setd2 KD-MA9 cells) compared to the control MA9 cells (Figure 3a). To further validate the crosstalk between SETD2-H3K36me3 and H3K79me2, we constructed a FLAG tagged vector, which contains all functional domains of SETD2, and named it as SETD2-Short (SETD2-S). Besides the elevation of H3K36me3, a dramatic reduction of H3K79me2 was observed by ectopic expression of SETD2 in MA9 cells (Figure 3b). Moreover, ectopic expression of SETD2 rescued H3K36me3 and reduced H3K79me2 in Setd2 KD-MA9 cells (Supplementary Figure S3b). To explore whether MLL fusion is necessary for the H3K79me2 increase, we performed Setd2 KD in WT HSPCs. Enhanced proliferation and self-renewal capacities were observed. A similar, but relatively mild, increase of H3K79me2 in Setd2 KDWT cells was detected (Supplementary Figures S3c and d). These data indicate that there is a crosstalk between SETD2 and H3K79me2, which is not MLL fusion specific, although MLL fusions may enhance this crosstalk.
Figure 3.
SETD2-H3K36me3 loss-of-function further elevated H3K79me2. (a) Immuno-blotting results after Setd2 KD in MA9 cells. (b) Schematic diagram showing major functional domains in SETD2 and the constructed SETD2-S vector. Immuno-blotting of SETD2-S and empty vector expression in MA9 cells. (c) ChIP-seq profiles of H3K79me2 and H3K36me3 for gene bodies (left: **P<0.01; right: ***P<0.001) in control (MA9_Control) and Setd2 KD-MA9 cells (MA9_shSetd2). (d) H3K79me2 and H3K36me3 ChIP signals on each gene. (e) Venn diagram of genes with altered H3K79me2 and/or H3K36me3 occupancies at gene bodies after Setd2 KD in MA9 cells. See also Supplementary Table S1.
To understand the crosstalk between H3K79me2 and H3K36me3 on genome-wide, ChIP-seq analysis was performed. No significant change was observed of peak distributions. However, a further increase of H3K79me2 was observed besides the expected decrease of H3K36me3 (Figure 3c, Supplementary Figures S4a and b). In MA9 HSPCs, H3K79me2 showed a preferential increase on MLL-AF9 targets but H3K36me3 showed a global increase (Figure 1c). To study whether the histone changes in Setd2 KD-MA9 cells are still different on MLL-AF9 targets and non-targets, we focused histone modification on these two groups of genes respectively. We found that the histone alteration patterns of H3K79me2 and H3K36me3 in Setd2 KD-MA9 cells were similar on MLL-AF9 targets or non-targets. Same results on MLL-ENL targets and non-targets were observed (Supplementary Figures S4c and d).
To explore the potential targets modulated by decreased H3K36me3 and further increased H3K79me2 by SETD2 LOF, we determined the genes with altered changes of these two marks in Setd2 KD-MA9 cells. Most of the genes with H3K79me2 and H3K36me3 alterations (8572/9064) showed decreased H3K36me3 and/or increased H3K79me2. We detected a significant overlap between genes with increased H3K79me2 and decreased H3K36me3, further supporting the co-enrichment of these two marks. We defined these genes as H3K79me2-H3K36me3 genes modulated by Setd2 KD (K79-K36-Setd2 genes) (Figures 3d and e, Supplementary Table S1).
Gene dysregulation in Setd2 KD-MA9 cells with decreased H3K36me3 and increased H3K79me2
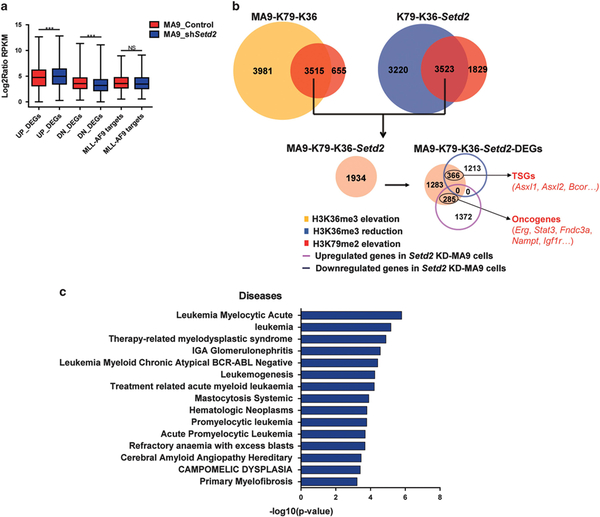
To identify the affected targets by Setd2 KD, we performed RNA-seq analysis of Setd2 KD-MA9 cells and control MA9 cells and identified significant differentially expressed genes (DEGs) (Supplementary Table S2). We found that both H3K79me2 and H3K36me3 showed dramatic changes on DEGs compared to non-DEGs (Supplementary Figure S5a), implicating the correlation between gene expression changes and histone modifications. Upregulation of MLL direct targets is a known key driver of MLLr leukemogenesis; thus we examined whether SETD2 inactivation accelerates leukemia through further activation of MLL-AF9 targets. Surprisingly, MLL-AF9 targets showed no significant expression change in Setd2 KD-MA9 cells (Figure 4a). The same comparison in Setd2 KD-WT cells versus control WT cells was conducted. No significant change was detected (Supplementary Figure S5b). These results indicate that the SETD2 LOF could mainly affect other genes but not known MLL-AF9 target genes.
Figure 4.
Gene dysregulation in Setd2 KD-MA9 cells with decreased H3K36me3 and increased H3K79me2. (a) Box plots showing that unlike upregulated genes and downregulated genes, there was no significant expression changes of selected MLL-AF9 targets in MA9_shSetd2 cells compared to MA9_Control cells. P-values were calculated using two-tailed t-test (***P<0.001; NS, P>0.05). (b) Workflow of using integrative ChIP-seq and RNA-seq analysis to identify MA9-K79-K36-Setd2-DEGs. Genes co-enriched with increased H3K79me2 and H3K36me3 in MA9 cells (MA9-K79-K36 genes), and also modulated by increased H3K79me2 and decreased H3K36me3 through Setd2 KD (K79-K36-Setd2 genes) were identified as MA9-K79-K36-Setd2 genes. AML-related TSGs and oncogenes are shown in red. See also Supplementary Tables S1–S3. (c) The MA9-K79-K36-Setd2-DEGs were mainly enriched in AML-related disease terms. Top 15 terms are shown according to P-value.
To identify the critical Setd2 KD affected genes which are modulated by H3K79me2 and H3K36me3 dynamic changes in MA9 cells, integrated analysis of ChIP-seq and RNA-seq was performed. Among the 3515 of MA9-K79-K36 genes co-enriched with increased H3K79me2 and H3K36me3 in MA9 HSPCs, 1934 of MA9-K79-K36-Setd2 genes were further modulated by increased H3K79me2 and decreased H3K36me3 through Setd2 KD. According to RNA-seq data, among the MA9-K79-K36-Setd2 genes, 366 of genes showed significant downregulation, whereas 285 of genes showed upregulation by Setd2 KD. We defined these DEGs as affected targets of Setd2 KD in MA9 cells modulated by dynamic H3K79me2 and H3K36me3 changes (MA9-K79-K36-Setd2-DEGs). MA9-K79-K36-Setd2-DEGs could be critical targets sensitive to Setd2 dosage and modulated by both H3K79me2 and H3K36me3 changes and contribute to cooperative leukemogenesis. Importantly, the TSGs and oncogenes co-enriched with increased H3K79me2 and H3K36me3 in MA9 cells were included in the MA9-K79-K36-Setd2-DEGs. The critical functions of these TSGs and oncogenes have been reported in leukemia patients18–24 (Figures 2c and 4b, Supplementary Tables S1–S3). Moreover, pathway analysis showed that AML-related disease terms were significantly enriched in MA9-K79-K36-Setd2-DEGs (Figure 4c), suggesting their functions in MLLr leukemogenesis. Our results suggest that H3K79me2 and H3K36me3 changes may accelerate AML through differential regulation of subsets of TSGs and oncogenes.
To understand whether the differential regulation on the subsets of TSGs and oncogenes is due to distinct histone modification changes, we calculated H3K79me2 and H3K36me3 ChIP signals on downregulated genes and upregulated genes respectively. Similar histone changes on both groups of genes were found (Supplementary Figure S5c). How this similar histone modification changes differentially affect gene regulation of TSGs and oncogenes is unclear.
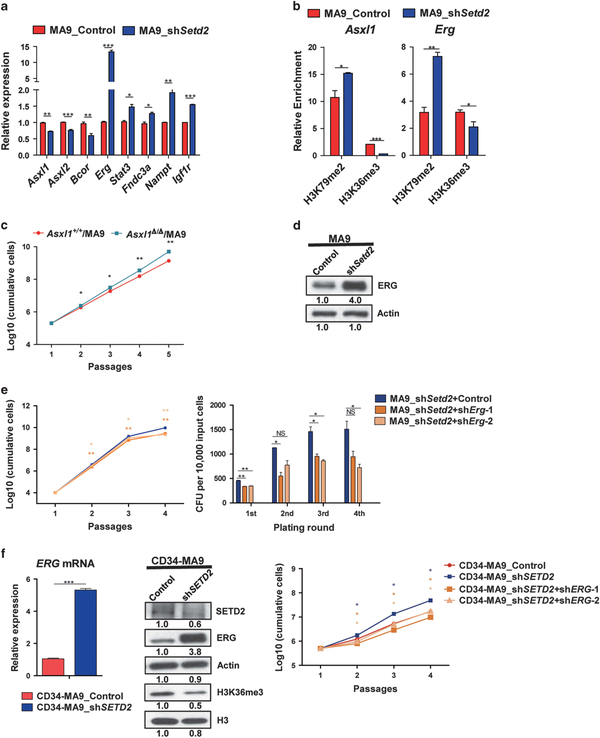
Both TSGs’ downregulation and oncogenes’ upregulation contribute to Setd2/SETD2 KD-mediated leukemia acceleration A set of TSGs and oncogenes were included in MA9-K79-K36-Setd2-DEGs (Figure 4b). To understand the function of these genes in leukemogenesis, we first validated their gene expression changes in Setd2 KD-MA9 cells. Significant downregulation of TSGs Asxl1, Asxl2, Bcor and upregulation of oncogenes Erg, Stat3, Fndc3a, Nampt, Igf1r genes were observed (Figure 5a). Increased H3K79me2 and decreased H3K36me3 were detected at these genes by ChIP-qPCR and ChIP-seq analysis (Figure 5b, Supplementary Figures S6a–c, S7a–S7e). Among the TSGs, the Asxl1 (Additional sex comb-like 1) gene is a member of the Enhancer of trithorax and Polycomb (ETP) genes. ASXL1 LOF mutations have been described in human myelodysplastic syndrome and AML patients including MLLr leukemia.25,26 Clinical studies have shown that ASXL1 mutations are unfavorable prognostic factors which indicate poor therapy outcomes for myelodysplastic syndrome and AML patients.27,28 To validate the TSG function of Asxl1 in MA9 cells, c-Kit+ cells from Asxl1 knock-out (Asxl1Δ/Δ) mouse were purified and transduced with MLL-AF9. Enhanced cell proliferation and enlarged colony size were observed in Asxl1Δ/Δ/MA9 cells compared to Asxl1+/+/MA9 cells. Unlike Setd2 KD, we did not observe significant enhancement of self-renewal capacity (Figure 5c, Supplementary Figures S8a andb), which indicates that Asxl1 enhanced cell proliferation more than leukemic stemness in MA9 cells.
Figure 5.
Both downregulation of Asxl1 and upregulation of Erg/ERG contribute to Setd2/SETD2 KD-mediated leukemia acceleration. (a) DEGs in Setd2 KD-MA9 cells were validated by RT-PCR. P-values were calculated by two-tailed Student’s t-test (*P<0.05, **P<0.01, ***P<0.001). Data are represented as mean ± s.d. (n = 2). Representative data are shown. (b) ChIP-qPCR of H3K79me2 and H3K36me3 at Asxl1 and Erg gene bodies. Enrichment was normalized to input samples (*P<0.05). Data are represented as mean ± s.d. (n = 2). (c) Enhanced proliferation of Asxl1Δ/Δ/MA9 cells compared to Asxl1+/+/MA9 cells was observed. P-values were calculated by two-tailed Student’s t-test (*P<0.05, **P<0.01). Data are represented as mean ± s.d. (n = 2). (d) Overexpression of ERG after Setd2 KD in MA9 cells was detected by immuno-blotting. (e) Decreased proliferation and self-renewal capacities in Erg KD MA9_shSetd2 cells (MA9_shSetd2+shErg) compared to control cells (MA9_shSetd2 +Control) (*P<0.05, **P<0.01). Data are represented as mean ± s.d. (n = 2). (f) Activation of ERG after SETD2 KD in human CD34-MA9 cells (CD34-MA9_shSETD2) compared to control cells (CD34-MA9_Control) (***P<0.001) was detected by RT-PCR and immuno-blotting. Enhanced cell proliferation of CD34-MA9_shSETD2 cells compared to CD34-MA9_Control cells was observed. ERG KD decreased cell proliferation in CD34-MA9_shSETD2 cells (CD34-MA9_shSETD2+shERG) compared to control cells (CD34-MA9_shSETD2) (*P<0.05).
Besides the repression of TSGs, Setd2 KD also leads to activation of oncogenes. Among the upregulated oncogenes, Erg is the most significantly activated one (Figure 5a). The ERG (Ets-related gene) belongs to the ETS-family transcription factors which plays an important role in normal hematopoiesis and leukemia. High expression of ERG is related to poor clinical outcomes in AML and ALL patients.29–31 Thus Erg upregulation by Setd2 KD could also be responsible for the acceleration of MLLr leukemogenesis. To validate this, we further explored Erg functions. Significant overexpression of Erg after Setd2 KD in MA9 cells was observed (Figure 5d). We also found upregulation of Erg on both mRNA and protein levels after Setd2 KD in WT cells, but the change was less profound compared to the MA9 cells (Supplementary Figures S8c and d). This indicates that the upregulation of Erg may be dependent on both Setd2 dosage and MLL fusion background. To validate the functional relevance of Erg in SETD2 LOF-MA9 leukemia, we further performed Erg KD in Setd2 KD-MA9 cells. Decreased self-renewal and proliferation capacities were found after Erg KD, which partially rescued the effects of Setd2 KD (Figure 5e, Supplementary Figures S8e and f). Moreover, significant activation of ERG by SETD2 KD was observed in human cord blood (CB) CD34+ cells derived MLL-AF9 cells (CB CD34+ cells were transduced with MLL-AF9 construct, CD34-MA9). ERG KD partially rescued the proliferation of SETD2 KD effect in CD34-MA9 cells (Figure 5f, Supplementary Figures S8g and h).
Furthermore, Asxl1 upregulation and Erg downregulation were observed by SETD2 ectopic expression in MA9 cells. We also detected Asxl1 upregulation and reversed Erg activation by SETD2 ectopic expression in Setd2 KD-MA9 cells, which rescued the Setd2 KD effects (Supplementary Figures S8i and j). These results support the key role of Asxl1 and Erg/ERG in MA9 leukemogenesis acceleration as Setd2/SETD2 targets modulated by H3K79me2 and H3K36me3 alterations.
DISCUSSION
In current study, using an MA9 genetic mouse model and through modulating the SETD2-H3K36me3 axis, we investigated the crosstalk between the DOT1L-H3K79me2 axis and the SETD2-H3K36me3 axis in MLLr leukemia under both SETD2-WT and SETD2 LOF contexts. Through integrated ChIP-seq and RNA-seq analysis, increased H3K79me2 and H3K36me3 were observed in SETD2-WT MLLr cells, and co-enrichment of histone changes in a large set of MA9-K79-K36 genes was identified. On mRNA expression levels, the upregulated genes are mainly MLL fusion targets, whereas subsets of additional TSGs and oncogenes showed no significant change but were epigenetically marked by these two histone modifications. By gain of second hit of SETD2 LOF (KD), decreased H3K36me3 and further increased HK379me2 were observed. These new changes did not further activate previously defined MLL fusion targets, but rather differentially regulated the previously marked subsets of TSGs and oncogenes. Downregulated TSGs including Asxl1 and upregulated oncogenes including Erg/ERG by Setd2/SETD2 KD were identified, showing the differential regulation of these two subsets of genes by Setd2/SETD2 which may partially explain the functional consequence of SETD2 LOF in MLLr leukemia (Supplementary Figure S9).
ChIP-seq data revealed increased H3K36me3, besides H3K79me2, at gene bodies in MA9 cells (Figure 1b, Supplementary Figure S1c), which may implicate that there is globally enhanced transcription elongation. The global increase of H3K36me3 is different from the increase of H3K79me2 which is selectively increased on MLL fusion targets (Figure 1c, Supplementary Figures S1d and e). Considering SETD2 is responsible for H3K36me3 modification, our results implicate that MLL fusions may activate SETD2 either directly or indirectly. This activation is not at specific gene loci but rather at the whole genome scale. Moreover, a large group of MA9-K79-K36 genes were identified which were co-enriched by H3K79me2 and H3K36me3 in MA9 cells (Figure 1e, Supplementary Table S1). Through correlation analysis and GSEA, we found that the gene dysregulation in MA9 is significantly correlated with increased H3K79me2 and H3K36me3. Many well-known MLL targets were included in the MA9-K79-K36 genes (Figures 2a and b). These results indicate that MLL-AF9 alters the downstream targets’ expression changes on limited genes with H3K79me2 changes. The increased H3K36me3 level mediated by SETD2 may also modulate MLL-AF9 targets in MLLr leukemia.
MA9 causes the upregulation of both H3K79me2 and H3K36me3, which suggests an activation of SETD2 function. Activation of oncogenic pathways could lead to activated tumor suppressor pathways. This MA9-SETD2 functional relationship may be another example of such a regulation loop, although it is unclear how SETD2-H3K36me3 is activated by MA9 either directly or indirectly. The high frequency of SETD2 mutations in MLLr leukemia indicated that disruption of this tumor suppressor pathway and the negative feedback loop is an ‘onco-requisition’ step for leukemia development. We confirmed that H3K36me3 was dramatically decreased after Setd2 KD; however, oncogenic DOT1L-H3K79me2 axis was further increased. Importantly, we found that ectopic expression of the functional SETD2 fragment could elevate H3K36me3 and reduce H3K79me2, which support the crosstalk between SETD2-H3K36me3 and H3K79me2 (Figure 3b and Supplementary Figure S3b). However, we did not observe further activation of MLL-AF9 targets by Setd2 KD as the expression levels of these targets were maintained at a relatively high level (Figure 4a). MA9-K79-K36-Setd2-DEGs sensitive to Setd2 dosage and modulated by both H3K79me2 and H3K36me3 changes were identified. Interestingly, these genes showed opposite gene expression changes after Setd2 KD. Downregulated genes include TSGs such as Asxl1; upregulated genes contain oncogenes such as Erg which have been reported playing key roles in hematopoiesis and/or leukemogenesis (Figure 4b, Supplementary Table S3).
Although reactivation of SETD2 or downstream TSGs, such as ASXL1, may be difficult, activated oncogenes may be easier to be targeted in leukemia therapy, as inhibitors are usually easier to identify than activators. We identified activation of Erg/ERG by Setd2/SETD2 KD in both mouse MA9 and human CD34-MA9 cells (Figures 5a,d,f). Also, Erg downregulation was observed by SETD2 ectopic expression in MA9 cells (Supplementary Figure S8i). ERG overexpression has been reported in variable human cancers such as prostate and sarcoma besides leukemia. High levels of ERG is a clinical biomarker of MLLr leukemia patients with poor outcomes.32–34 In our work, we found Erg/ERG KD could rescue the enhanced proliferation and self-renewal advantage by Setd2/SETD2 KD (Figures 5e and f), indicating the important function of ERG as a target of SETD2 in MLLr leukemia acceleration. This also implicates that ERG could be a novel therapeutic target in leukemia patients with SETD2 mutations.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by the National Natural Science Foundation of China (grant 81425003 and 91731302 to QFW, grant 81500142 to AC), the Strategic Priority Research Program of the Chinese Academy of Sciences (grant XDA12010304 to QFW), the CFK (to GH), National Institutes of Health (NIH) (R21CA187276 to GH), the scholarship from China Scholarship Council (No. 201504910521 to JB), the Youth Innovation Promotion Association of Chinese Academy of Sciences (to FH), the Suzhou Key Laboratory for Pediatric Leukemia (SZS201615), Social Development Project of Jiangsu Province (CXTDA2017014 to SH), the Uehara Memorial Foundation for a Research Fellowship (to YH). We thank Dr Tao Cheng (Chinese Academy of Medical Sciences and Peking Union Medical College) for generously providing the Setd2 shRNAs.
Footnotes
Supplementary Information accompanies this paper on the Leukemia website (http://www.nature.com/leu)
CONFLICT OF INTEREST
The authors declare no conflict of interest.
REFERENCES
- 1.Wagner EJ, Carpenter PB. Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 2012; 13: 115–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalgliesh GL, Furge K, Greenman C, Chen L, Bignell G, Butler A et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature 2010; 463: 360–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Duns G, van den Berg E, van Duivenbode I, Osinga J, Hollema H, Hofstra RMW et al. Histone methyltransferase gene SETD2 is a novel tumor suppressor gene in clear cell renal cell carcinoma. Cancer Res 2010; 70: 4287–4291. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Duns G, Westers H, Sijmons R, van den Berg A, Kok K. SETD2: an epigenetic modifier with tumor suppressor functionality. Oncotarget 2016; 7: 50719–50734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhu X, He F, Zeng H, Ling S, Chen A, Wang Y et al. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat Genet 2014; 46: 287–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mar BG, Bullinger LB, McLean KM, Grauman PV, Harris MH, Stevenson K et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat Commun 2014; 5: 3469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersson AK, Ma J, Wang J, Chen X, Gedman AL, Dang J et al. The landscape of somatic mutations in infant MLL-rearranged acute lymphoblastic leukemias. Nat Genet 2015; 47: 330–337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J Biol Chem 2002; 277: 49383–49388. [DOI] [PubMed] [Google Scholar]
- 9.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol Cell Biol 2005; 25: 3305–3316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bannister AJ, Schneider R, Myers FA, Thorne AW, Crane-Robinson C, Kouzarides T. Spatial distribution of di- and tri-methyl lysine 36 of histone H3 at active genes. J Biol Chem 2005; 280: 17732–17736. [DOI] [PubMed] [Google Scholar]
- 11.Guenther MG, Levine SS, Boyer LA, Jaenisch R, Young RA. A chromatin landmark and transcription initiation at most promoters in human cells. Cell 2007; 130: 77–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonkers I, Lis JT. Getting up to speed with transcription elongation by RNA polymerase II. Nat Rev Mol Cell Biol 2015; 16: 167–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Steger DJ, Lefterova MI, Ying L, Stonestrom AJ, Schupp M, Zhuo D et al. DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol Cell Biol 2008; 28: 2825–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang QF, Wu G, Mi S, He F, Wu J, Dong J et al. MLL fusion proteins preferentially regulate a subset of wild-type MLL target genes in the leukemic genome. Blood 2011; 117: 6895–6905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bernt KM, Zhu N, Sinha AU, Vempati S, Faber J, Krivtsov AV et al. MLL-rearranged leukemia is dependent on aberrant H3K79 methylation by DOT1L. Cancer Cell 2011; 20: 66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Okada Y, Feng Q, Lin Y, Jiang Q, Li Y, Coffield VM et al. hDOT1L links histone methylation to leukemogenesis. Cell 2005; 121: 167–178. [DOI] [PubMed] [Google Scholar]
- 17.Deshpande AJ, Deshpande A, Sinha AU, Chen L, Chang J, Cihan A et al. AF10 regulates progressive H3K79 methylation and HOX gene expression in diverse AML subtypes. Cancer Cell 2014; 26: 896–908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gelsi-Boyer V, Trouplin V, Adelaide J, Bonansea J, Cervera N, Carbuccia N et al. Mutations of polycomb-associated gene ASXL1 in myelodysplastic syndromes and chronic myelomonocytic leukaemia. Br J Haematol 2009; 145: 788–800. [DOI] [PubMed] [Google Scholar]
- 19.Micol JB, Duployez N, Boissel N, Petit A, Geffroy S, Nibourel O et al. Frequent ASXL2 mutations in acute myeloid leukemia patients with t(8;21)/RUNX1-RUNX1T1 chromosomal translocations. Blood 2014; 124: 1445–1449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Grossmann V, Tiacci E, Holmes AB, Kohlmann A, Martelli MP, Kern W et al. Whole-exome sequencing identifies somatic mutations of BCOR in acute myeloid leukemia with normal karyotype. Blood 2011; 118: 6153–6163. [DOI] [PubMed] [Google Scholar]
- 21.Tsuzuki S, Taguchi O, Seto M. Promotion and maintenance of leukemia by ERG. Blood 2011; 117: 3858–3868. [DOI] [PubMed] [Google Scholar]
- 22.Koskela HL, Eldfors S, Ellonen P, van Adrichem AJ, Kuusanmaki H, Andersson EI et al. Somatic STAT3 mutations in large granular lymphocytic leukemia. N Engl J Med 2012; 366: 1905–1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Audrito V, Serra S, Brusa D, Mazzola F, Arruga F, Vaisitti T et al. Extracellular nicotinamide phosphoribosyltransferase (NAMPT) promotes M2 macrophage polarization in chronic lymphocytic leukemia. Blood 2015; 125: 111–123. [DOI] [PubMed] [Google Scholar]
- 24.Medyouf H, Gusscott S, Wang H, Tseng JC, Wai C, Nemirovsky O et al. High-level IGF1R expression is required for leukemia-initiating cell activity in T-ALL and is supported by Notch signaling. J Exp Med 2011; 208: 1809–1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lavallee VP, Baccelli I, Krosl J, Wilhelm B, Barabe F, Gendron P et al. The transcriptomic landscape and directed chemical interrogation of MLL-rearranged acute myeloid leukemias. Nat Genet 2015; 47: 1030–1037. [DOI] [PubMed] [Google Scholar]
- 26.Patel JP, Gonen M, Figueroa ME, Fernandez H, Sun Z, Racevskis J et al. Prognostic relevance of integrated genetic profiling in acute myeloid leukemia. N Engl J Med 2012; 366: 1079–1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bejar R, Stevenson K, Abdel-Wahab O, Galili N, Nilsson B, Garcia-Manero G et al. Clinical effect of point mutations in myelodysplastic syndromes. N Engl J Med 2011; 364: 2496–2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abdel-Wahab O, Adli M, LaFave LM, Gao J, Hricik T, Shih AH et al. ASXL1 mutations promote myeloid transformation through loss of PRC2-mediated gene repression. Cancer Cell 2012; 22: 180–193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baldus CD, Burmeister T, Martus P, Schwartz S, Gokbuget N, Bloomfield CD et al. High expression of the ETS transcription factor ERG predicts adverse outcome in acute T-lymphoblastic leukemia in adults. J Clin Oncol 2006; 24: 4714–4720. [DOI] [PubMed] [Google Scholar]
- 30.Thoms JA, Birger Y, Foster S, Knezevic K, Kirschenbaum Y, Chandrakanthan V et al. ERG promotes T-acute lymphoblastic leukemia and is transcriptionally regulated in leukemic cells by a stem cell enhancer. Blood 2011; 117: 7079–7089. [DOI] [PubMed] [Google Scholar]
- 31.Tursky ML, Beck D, Thoms JA, Huang Y, Kumari A, Unnikrishnan A et al. Over-expression of ERG in cord blood progenitors promotes expansion and recapitulates molecular signatures of high ERG leukemias. Leukemia 2015; 29: 819–827. [DOI] [PubMed] [Google Scholar]
- 32.Loughran SJ, Kruse EA, Hacking DF, de Graaf CA, Hyland CD, Willson TA et al. The transcription factor Erg is essential for definitive hematopoiesis and the function of adult hematopoietic stem cells. Nat Immunol 2008; 9: 810–819. [DOI] [PubMed] [Google Scholar]
- 33.Stankiewicz MJ, Crispino JD. ETS2 and ERG promote megakaryopoiesis and synergize with alterations in GATA-1 to immortalize hematopoietic progenitor cells. Blood 2009; 113: 3337–3347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pereira DS, Dorrell C, Ito CY, Gan OI, Murdoch B, Rao VN et al. Retroviral transduction of TLS-ERG initiates a leukemogenic program in normal human hematopoietic cells. Proc Natl Acad Sci USA 1998; 95: 8239–8244. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.