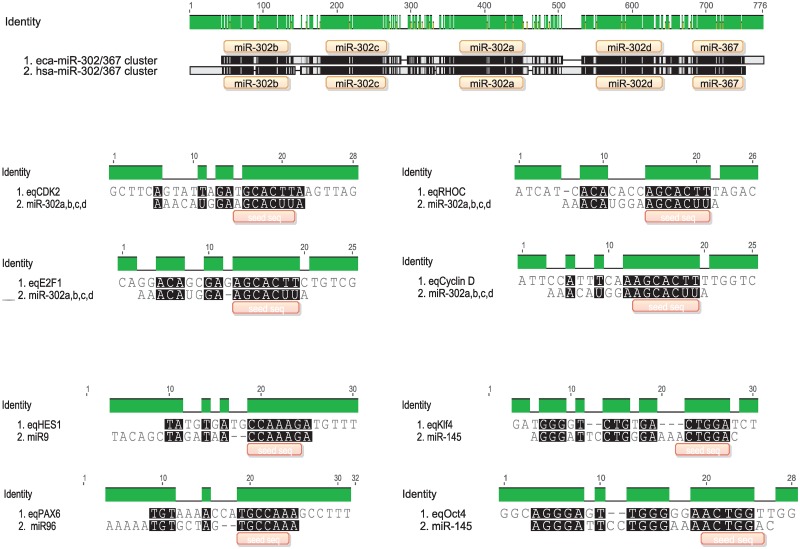
Fig 6. In-silico analysis of equine microRNAs targets.
The green bars and the black zones represent the homology between sequences. a) Comparison between equine miR-302/367 cluster (eca-miR-302/367) and human miR-302/367 cluster (hsa-miR-302/367) in the genome. In orange are the different microRNAs positioned in the cluster. Below this alignment, the seed sequence of the miR-302 family is aligned to the 3‘UTR of 4 mRNAs in the equine (CDK2, E2F1, RHOC and CYCLIN D). b) Seed alignment of the differentiation-related microRNAs miR-9, miR-96 and miR-145, to the 3‘UTR of the equine mRNAs HES1, PAX6 and KLF4 and OCT-4, respectively.