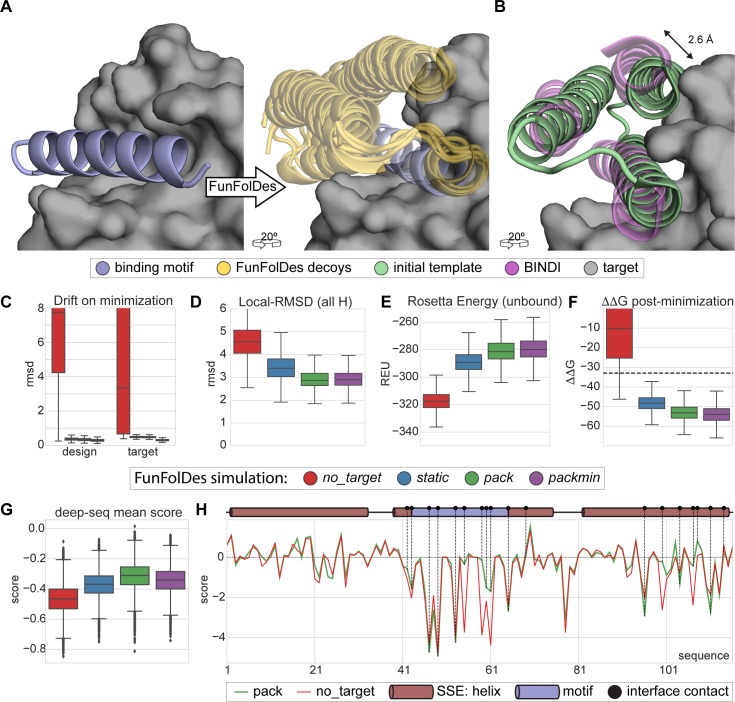
Fig 4. Target-biased design of a protein binder and performance assessment based on saturation mutagenesis.
A) Depiction of the initial design task, a single-segment binding motif (BIM-BH3) shown in light blue cartoons, with its target (BHRF1) shown in gray surface, is used by FunFolDes to generate an ensemble of designs compatible with the binding mode shown in light orange cartoons. B) Conformational difference between the initial template (PDB ID: 3LHP), shown in light brown and the previously designed binder (BINDI), shown in violet cartoons, helix 3 requires a subtle but necessary shift (2.6 Å) to avoid steric clashes with the target. C-G) Scoring metrics for design populations according to the simulation mode: no_target—FunFolDes was used without the target protein; static—target present no flexibility allowed; pack—target allowed to repack the side-chains; packmin–side-chain repacking plus minimization and backbone minimization were allowed for the target. The target flexibility was allowed during the relax-design cycles of FunFolDes. C) Structural drift observed for design and target binder measured as the RMSD between pre- and post-minimization conformations. D) Structural recovery of the conformation observed in the BINDI-BHRF1 assessed over the 3 helical segments of the bundle. E) Rosetta energy for the designs in the unbound state generated by different simulation modes. F) Interaction energy (ΔΔG) between the designs and the target. G) Deep-sequencing score distribution for each design population, computed as the mean score of each sequence after applying a position score matrix based on the deep-sequencing data. The pack population slightly outperforms the other simulation modes. H) Per-residue scoring comparison of the no_target and the pack populations according to the deep-sequencing data. Although the behavior is overall similar, pack outperforms no_target in multiple positions, several of which are highlighted (black dots) as interfacial contacts or second shell residues close to the binding site.