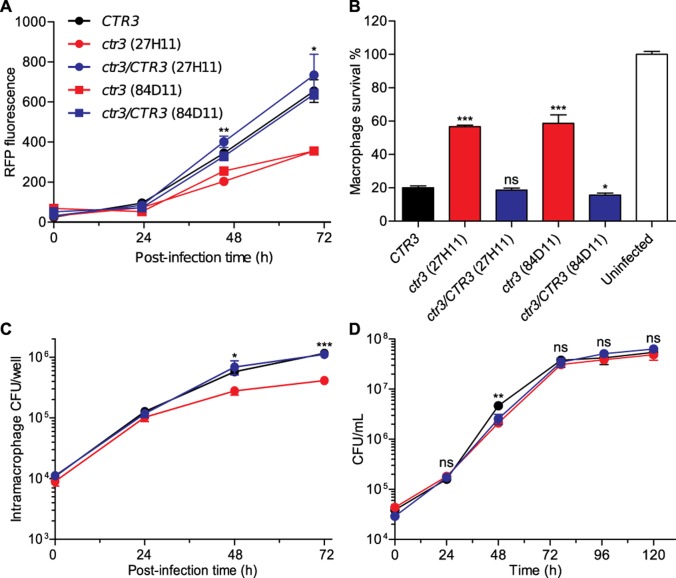
Fig 1. H. capsulatum intramacrophage growth requires Ctr3.
H. capsulatum proliferation in macrophages was measured by fluorescence of RFP-expressing yeasts (A), yeast-growth dependent lysis of the macrophage population (B), and determination of intracellular viable yeasts (C). (A) RFP-fluorescence of intracellular H. capsulatum yeasts was monitored over 72 hours following infection of P388D1 macrophages at an MOI of 1:2 with CTR3-expressing yeasts (CTR3; black), two mutants with the CTR3 gene disrupted by T-DNA insertion (ctr3 27H11 (circles) and 84D11(squares); red), or mutant strains complemented with a wild-type CTR3 gene (ctr3/CTR3; blue). (B) Survival of lacZ-expressing P388D1 macrophages was quantified after 7 days of infection by removal of culture medium and measurement of the remaining macrophage-derived β-galactosidase activity. (C) Viability of intracellular H. capsulatum yeasts over 72 hours was determined by lysing infected macrophages and plating of the lysate on solid HMM medium to enumerate colony forming units (CFU). (D) Growth of CTR3-expressing (black), ctr3 mutant (27H11; red), and the ctr3/CTR3 complemented (blue) strains in liquid culture in HMM at 37°C was measured by plating dilutions of the culture on solid medium to enumerate CFU. All data represent the average ± standard deviation of biological replicates (n = 3). Statistically significant differences between CTR3 and ctr3 strains were determined by one-tailed Student’s t-test and are indicated with asterisks (* P < 0.05, ** P < 0.01, *** P < 0.001).