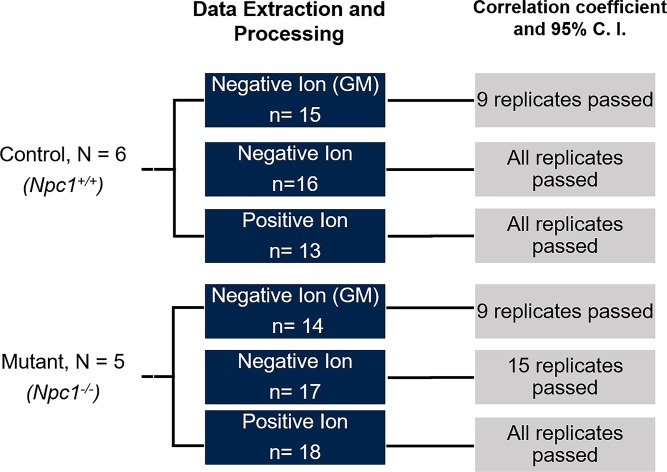
Fig. 4.
General workflow for collection and processing of multiple biological and technical replicates in MALDI-MSI datasets. Biological replicates were processed by investigating n = 6 (control) and n = 5 (mutant) animals. Cerebellar tissue was imaged using MALDI-MSI, and spectra were collected in three experiments to detect GM, negative, and positive lipid ions. Using technical replicates from each animal, as indicated by “n,” the newly developed alignment, normalization, and consensus spectra algorithm was utilized. To evaluate spectral reproducibility, a dot product analysis was carried out and evaluated based on correlation. Outlier spectra were removed, and those that “passed” are indicated by having a correlation above the 95% CI.