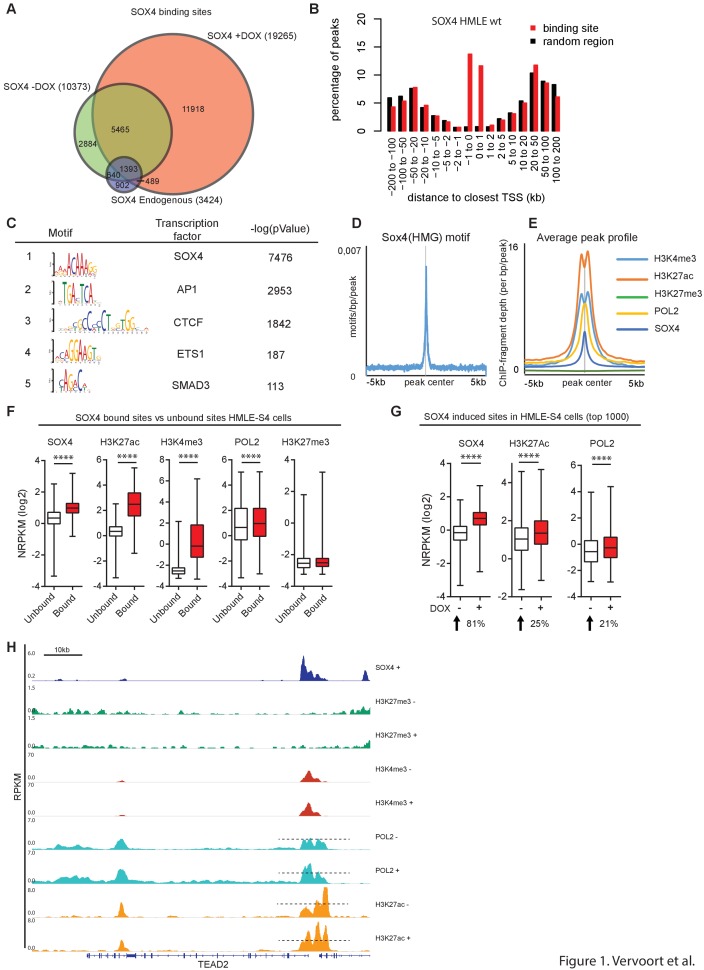
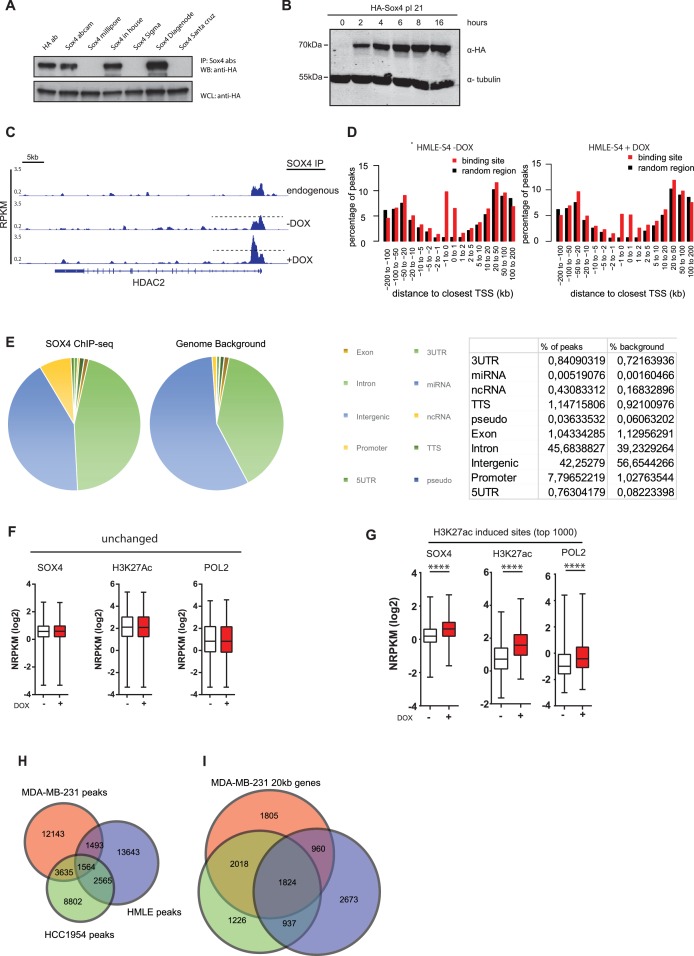
Figure 1. SOX4 is a transcriptional activator, preferentially binding to open/active chromatin.
(A) Venn-diagram showing the overlap of SOX4 binding sites identified by ChIP-seq in WT-HMLE and DOX-induced HMLE-S4 cells. (B). Genomic distribution of SOX4 binding sites in WT-HMLE cells with respect to annotated genes, represented as distance from the nearest TSS. Random genomic regions were used as background. (C). De novo motif analysis of SOX4-bound sites in DOX-treated HMLE-S4 cells. Selected significantly enriched motifs are represented. SOX4 was identified as the most highly enriched motif. (D) Density of the consensus SOX4 motif in SOX4 bound sites in HMLE-S4 cells 500 bp up- and downstream of the peak center. (E) Occupancy plots of SOX4, H3K27ac, H3K27me3, POL2 and H3K4me3 in the 10kb-genomic region surrounding the SOX4 peak center. (F) Changes in SOX4, H3K27ac, H3K4me3, POL2 and H3K27me3 are shown for SOX4-bound and SOX4-unbound sites (5%–95% whiskers, two-tailed Mann-Whitney U test, **** indicate p<0.0001). (G) Changes in SOX4, H3K27ac, and POL2 in HMLE-S4 for top 1000 DOX-induced sites ranked by SOX4-signal (****p<0.0001, Wilcoxon-signed-rank test) (H) Genomic tracks representing the occupancy of SOX4, H3K27me3, H3K4me3, H3K27ac and POL2 in untreated and DOX-treated HMLE-S4 cells surrounding the genomic locus of the canonical SOX4 target gene TEAD2.