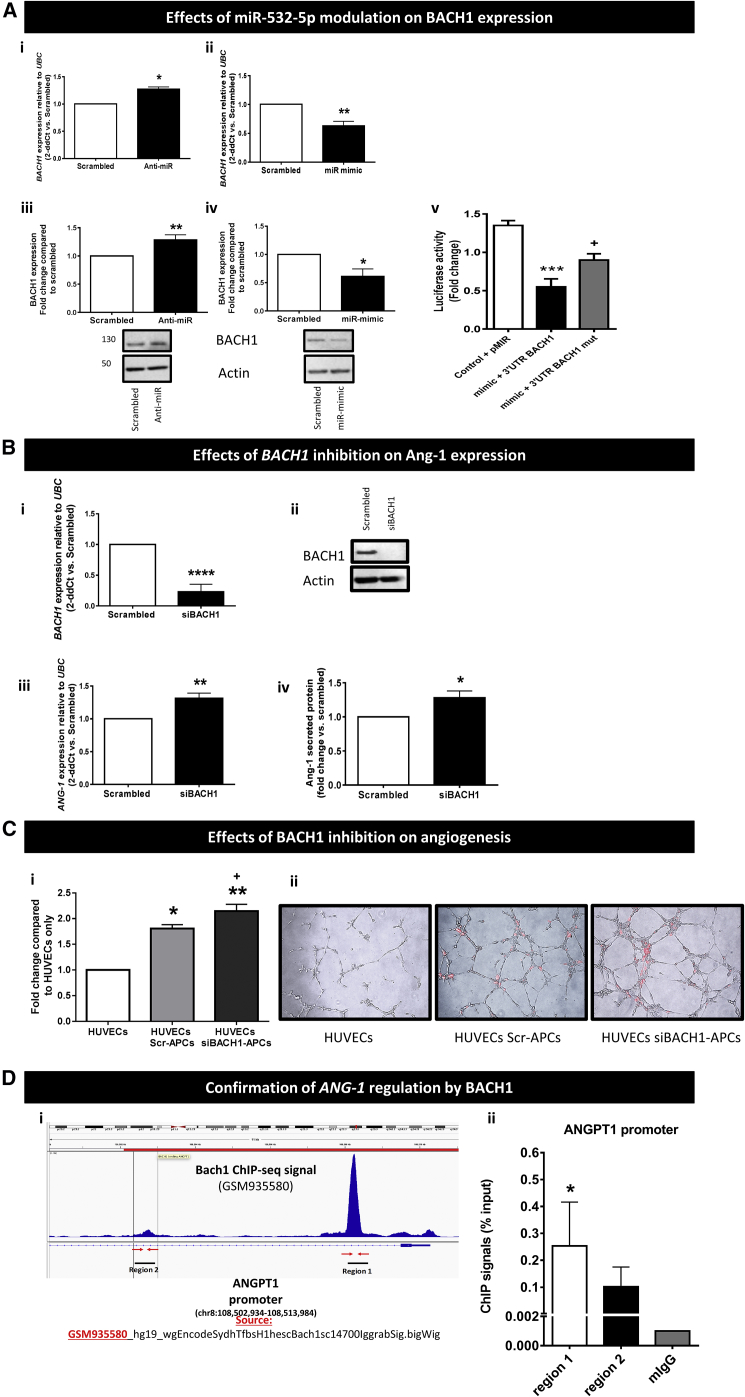
Figure 6.
miR-532-5p Regulates ANG-1 via Inhibition of the Transcription Factor BACH1
(A) qPCR (i and ii) and western blotting (iii and iv) demonstrating the effect of miR-532-5p inhibition (i and iii) (n = 4 biological replicates) and overexpression (ii and iv) (n = 5 biological replicates) on BACH1 gene and protein expression in APCs. *p < 0.05 and **p < 0.01 versus scrambled. v, Luciferase activity at 48 hr post-co-transfection of HEK293T cells with miR-532-5p mimics and the following plasmids: pMIR reported (empty vector), 3′ UTR-BACH1, and 3′ UTR-BACH1mut (with mutation of the putative miRNA target site) (n = 5 replicates). ***p < 0.001 versus control, +p < 0.05 versus mimic 3′ UTR BACH1. (B) qPCR, n = 4 (i), and representative western blot image, n = 3 (ii), demonstrating siRNA silenced both BACH1 gene (****p < 0.0001 versus control) and protein expression, respectively. Silencing BACH1 resulted in an increase ANG-1 mRNA (n = 6) (iii) and Ang-1 protein (n = 5) (iv). *p < 0.05 and **p < 0.01 versus scrambled. (C) Inhibition of BACH1 expression produced a significant increase in network formation compared with control (i); representative images, red color: DiL staining of pericytes, magnification ×10. n = 7 biological replicates (ii), *p < 0.05 and **p < 0.01 versus HUVECs, +p < 0.05 versus HUVECs + Scr-APCs. All values are means ± SE. (D) (i) BACH1 transcription factor binding sites by ENCODE ChIP-seq from NCBI GEO: GSM935580 were consulted and subsequent ChIP-qPCR performed for BACH1 occupancy of the ANGPT1 promoter regions (1) chr8:108,507,212–108,509,973 and (2) chr8:108,499,983–108,505,507, transcribed in [−] direction. (ii) ChIP-qPCR analysis confirms the regulation of ANG-1 by BACH1. qPCR values are expressed as mean ± SE and are representative of n = 3 independently performed qPCR experiments with one biological replicate of APC. *p < 0.05 versus mIgG.