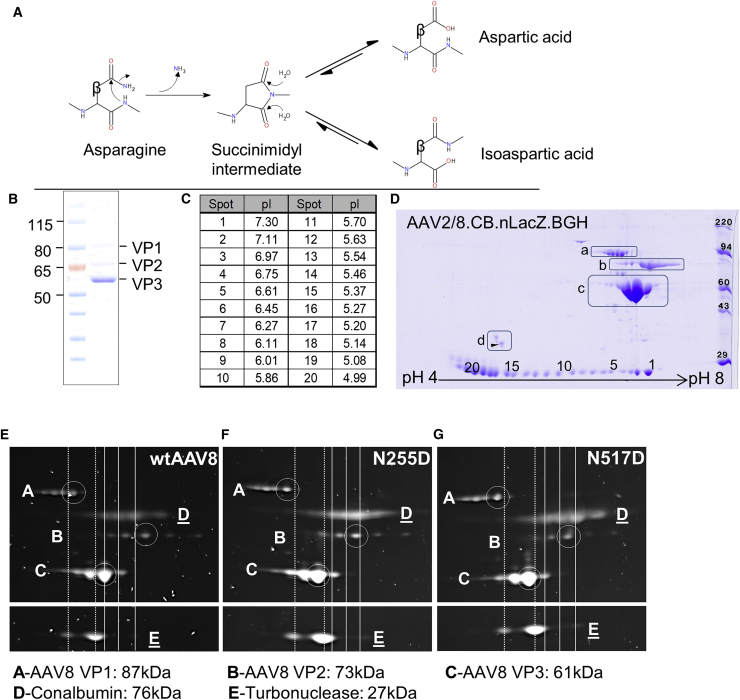
Figure 1.
Electrophoretic Analysis of AAV8 VP Isoforms
(A) Diagram illustrating the mechanism by which asparagine residues undergo nucleophilic attack by adjacent nitrogen atoms, forming a succinimidyl intermediate. This intermediate then undergoes hydrolysis, resolving into a mixture of aspartic acid and isoaspartic acid. The beta carbon is labeled as such. The diagram was generated in BIOVIA Draw 2018. (B) 1 μg of AAV8 vector was run on a denaturing one-dimensional SDS-PAGE. (C) Isoelectric points of carbonic anhydrase pI marker spots are shown. (D) 5 μg of AAV8 vector was analyzed by two-dimensional gel electrophoresis and stained with Coomassie blue. Spots 1–20 are carbamylated carbonic anhydrase pI markers. Boxed regions are as follows: a, VP1; b, VP2; c, VP3; and d, internal tropomyosin marker (arrow: tropomyosin spot of molecular weight [MW] = 33kDa, pI = 5.2). Isoelectric focusing was performed with a pI range of 4–8. 1 × 1011 GC of WT AAV8 (E) or mutant (F, N255D; and G, N517D) vector were analyzed by 2D gel electrophoresis and stained with SYPRO Ruby. Protein labeling: A, VP1; B, VP2; C, VP3; D, chicken egg white conalbumin marker; E, turbonuclease marker. Isoelectric focusing was performed with a pI range of 6–10. Primary VP1/2/3 isoform spots are circled, and migration distances of major spots of markers are indicated by vertical lines (turbonuclease, dashed; conalbumin, solid).