Figure 3.
Structural Modeling of the AAV8 VP3 Monomer and Analysis of Deamidated Sites
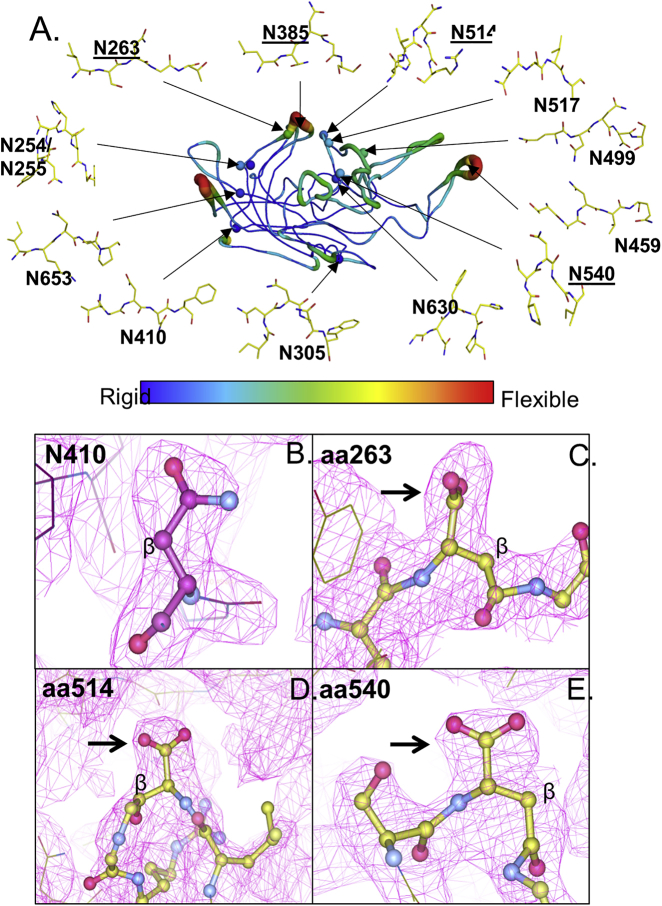
(A) The AAV8 VP3 monomer (PDB: 3RA8) is shown in a coil representation. The color of the ribbon indicates the relative degree of flexibility (blue = most rigid/normal temperature factor; red = most flexible/high temperature factor). Spheres indicate residues of interest. Expanded diagrams are ball-and-stick representations of residues of interest and their surrounding residues to demonstrate local protein structure (blue, nitrogen; red, oxygen). Underlined residues are those in NG motifs. (B–E) Isoaspartic models of deamidated asparagines with N+1 glycines are shown. The 2FoFc electron density map (1 sigma level) generated from refinement of the AAV8 crystal structure (PDB: 3RA8) with (B) an asparagine model of N410 in comparison with isoaspartic acid models of (C) N263, (D) N514, and (E) N540. Electron density map is shown in the magenta grid. All atoms are colored by atom type: carbon, purple (B)/gree6n (C–E); nitrogen, blue; oxygen, pink. The beta carbon is labeled as such. The arrows indicate electron density corresponding to the R group of the residue of interest.