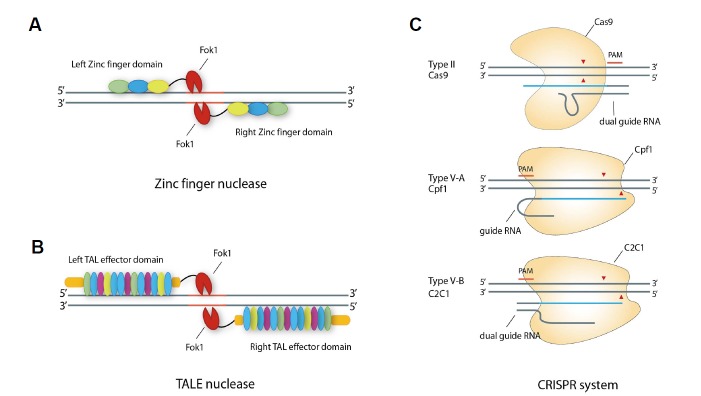
Fig. 3. Reprogrammable tools for target DNA-specific engineering.
(A) First-generation genome engineering system: ZFN. The zinc finger module, which specifically binds the target DNA sequence, is flanked by the type II endonuclease FokI. ZFN is activated when FokI endonuclease is dimerized by left and right pair-wise binding of the zinc finger motif at the target genomic sites. Ellipses indicate Zinc finger-binding modules, orange bold lines indicate the target sequence. (B) Second-generation genome engineering system: TALEN. The TAL effector (TALE) DNA-binding module is flanked by type II endonuclease FokI. TALEN requires dimerization to cleave the target DNA sequence. Within TALEN, DNA-binding specificity can be adjusted at the single-nucleotide level. Ellipses indicate TALEN-binding modules. Orange bold line on the DNA indicates the target sequence. (C) Third-generation genome engineering system: Class (II) CRISPR endonuclease. CRISPR is composed of effector protein and guide RNA with sequences complementary to the target DNA. Red and blue bold lines indicate the protospacer adjacent motif and target sequence, respectively, and the arrowhead indicates the DNA cleavage site.