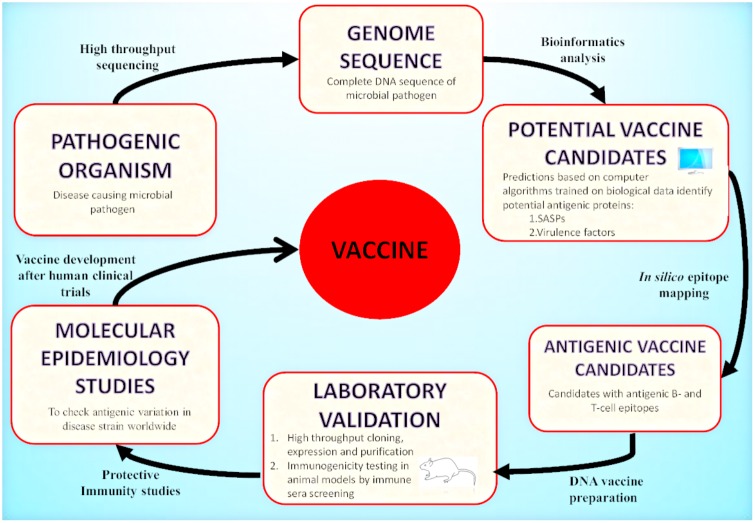
FIGURE 2.
Reverse Vaccinology approach: A schematic representation of vaccine development by RV is illustrated in the presented flowchart. RV starts with the computational analysis of the complete genome sequence of the targeted pathogenic organism. Computational predictions are based on algorithms trained on biological data obtained from experimentally carried out studies. The potential vaccine candidates include surface associated and secretory proteins (SASPs) and virulence factors. These are further evaluated to identify protein candidates with antigenic epitopes for B-cells and T-cells. These proteins are then amplified by PCR and expressed in suitable vectors. The recombinant proteins produced are purified and used for immunogenicity testing in animal models (mice). Based on immune sera screening (FACs, Serum Bactericidal Activity), the recombinant proteins capable of inducing sera bactericidal antibodies are selected. The top candidates enter the pre-clinical stage of vaccine development. After the molecular epidemiological studies, the best candidates are used for clinical trials in adults, adolescents and infants and finally they enter the vaccine formulation process.