Abstract
Signaling by classic analgesics, such as morphine, is governed primarily by the relative abundance of opioid receptors at the cell surface, and this is regulated by receptor delivery to, and retrieval from, the plasma membrane. Although retrieval mechanisms, such as receptor endocytosis, have been extensively investigated, fewer studies have explored mechanisms of receptor maturation and delivery to the plasma membrane. A previous study implicated receptor transporter proteins (RTPs) in the latter process. Since not much is known about regulation of RTP expression, we initiated studies examining the effect of chronic morphine administration on the levels of RTPs in the brain. Among the four RTPs, we detected selective and region-specific changes in RTP4 expression; RTP4 mRNA is significantly upregulated in the hypothalamus compared with other brain regions. We examined whether increased RTP4 expression impacted receptor protein levels and found a significant increase in the abundance of mu opioid receptors (MOPrs) but not other related G protein–coupled receptors (GPCRs, such as delta opioid, CB1 cannabinoid, or D2 dopamine receptors) in hypothalamic membranes from animals chronically treated with morphine. Next, we used a cell culture system to show that RTP4 expression is necessary and sufficient for regulating opioid receptor abundance at the cell surface. Interestingly, selective MOPr-mediated increase in RTP4 expression leads to increases in cell surface levels of MOPr–delta opioid receptor heteromers, and this increase is significantly attenuated by RTP4 small interfering RNA. Together, these results suggest that RTP4 expression is regulated by chronic morphine administration, and this, in turn, regulates opioid receptor cell surface levels and function.
Introduction
G protein–coupled receptors (GPCRs) belong to the seven transmembrane receptor family and the majority localize to the cell surface. The levels of GPCRs at the cell surface determine the extent of agonist-induced cellular responses by peptide and other hydrophylic ligands. Activation of these receptors leads to initiation of signaling via diverse signal-transduction pathways, including G protein- and β-arrestin–mediated signaling pathways, leading to their multiple physiologic effects.
Although significant effort has been put toward understanding mechanisms regulating receptor endocytosis due to its role in cellular desensitization [see review by Williams et al. (2013)], relatively few studies have focused on exploring mechanisms regulating delivery of GPCRs to the plasma membranes. Some of these studies suggested that chaperone proteins could regulate the glycosylation of GPCRs, an important step for membrane trafficking of receptors (Katada et al., 2004). Other studies have suggested a requirement of chaperone proteins for appropriate targeting to the cell surface membrane (Achour et al., 2008; Williams and Devi, 2010), such as NinaA for Drosophila rhodopsin, RanBP2 for mammalian cone opsin, ODR-4 for Caenorhabditis elegans chemosensory receptors, Receptor-Activity Modifying Proteins (RAMPs) for the mammalian calcitonin receptor like receptor, and receptor expression–enhancing proteins and receptor transporter proteins (RTPs) for the mammalian odorant and taste receptors (Saito et al., 2004; Behrens et al., 2006; Achour et al., 2008; Matsunami et al., 2009). In some cases, the chaperone proteins mask the endoplasmic reticulum retention signal present in GPCRs, thereby promoting cell surface receptor expression (Saito et al., 2004). These observations suggest that chaperone proteins play a crucial role in the maturation of GPCRs. However, very little information is available about how the expression or function of these chaperone proteins is regulated. In this study, we focused on RTPs and explored a role for their regulation in opioid receptor function.
Opioid receptors, members of class A GPCRs, bind to and are activated by opiates such as morphine and endogenous opioid peptides such as endorphins, enkephalins, and dynorphins. Studies by several groups, including ours, revealed that mu opioid receptors (MOPrs) and delta opioid receptors (DOPrs) form heteromers that exhibit pharmacological profiles that are distinct from the individual receptor protomers (Gomes et al., 2000, 2011; Rozenfeld and Devi, 2007). We have previously reported that RTP4, a member of the RTP family, plays an important role in the cell surface expression of these receptors. Furthermore, we showed that RTP4 facilitates cell surface expression of the heteromers; coexpression of RTP4 with MOPr and DOPr leads to enhanced cell surface expression as well as decreased ubiquitination of the receptors; and in the absence of RTP4, there is a specific retention of MOPr in the Golgi compartment, resulting in decreased cell surface expression of both protomers (Décaillot et al., 2008). These results imply that RTP4 regulates the membrane expression of not only MOPr and DOPr but also MOPr-DOPr heteromers, thereby playing an important role in influencing the action of exogenous and endogenous opioid ligands. In the context of opioid receptor ligands, we previously observed that chronic morphine administration can upregulate the expression of MOPr-DOPr heteromers in brain regions that are important for pain perception, as detected by using a MOPr-DOPr heteromer-selective antibody (Gupta et al., 2010). However, the mechanism of upregulation of MOPr-DOPr heteromers in these brain regions is not clear. More importantly, nothing is known about the mechanisms regulating RTP4 expression in general or by opioid receptor activation in particular. In this study, we examined the effect of morphine administration on RTP4 levels and the contribution of RTP4 to changes in cell surface expression of MOPr-DOPr heteromers in vitro and in vivo.
Materials and Methods
Cell Culture and Transfection.
Neuro 2A cells were grown in complete growth medium (Eagle’s Minimum Essential Medium (E-MEM) with 10% fetal bovine serum and 1% Penicillin Streptomycin (P/S)). Cells were transfected with either Flag-MOPr (N2AMOPr cells) or Flag-MOPr along with RTP4 small interfering RNA (siRNA; Sigma-Aldrich, St. Louis, MO) using Lipofectamine 2000 according to the manufacturer’s protocol (Thermo Fisher Scientific, Waltham, MA). Twenty-four hours after transfection, cells were seeded into 24-well plates, 96-well plates, or a LabTek chamber (Thermo Fisher Scientific) for further experiments.
Measurement of Gαi Response.
Gαi response was measured by using the CellKey System (Molecular Devices, Sunnyvale, CA), based on a label-free technology called cellular dielectric spectroscopy, which is capable of measuring complex impedance changes in cell monolayers after GPCR activation (an increase in impedance means Gαi activation) as described previously (Zhou et al., 2013). In brief, naïve or transiently transfected Neuro 2A cells (N2AMOPr cells) (32,000 cells/well) were seeded onto a CellKey poly-d-lysine–coated 96-well microplate (Molecular Devices) according to their optimum growth conditions on the day prior to the experiment and incubated overnight at 37°C in 5% CO2. At 30–60 minutes prior to running the assay, cells were washed with 150 µl of assay buffer (Hanks’ balanced salt solution with 20 mM HEPES and 0.1% bovine serum albumin, prewarmed at 29°C) followed by the addition of 135 µl of the assay buffer. The baseline impedance was measured for 2 minutes followed by the addition of ligands at a final concentration of 1 or 10 µM. The changes in impedance were measured for 20 minutes after addition of ligands.
Treatment of N2AMOPr Cells.
Twenty-four hours after plasmid and/or siRNA transfection, N2AMOPr cells were seeded into a 24-well plate (200,000 cells/well) in complete growth medium and incubated overnight at 37°C with 5% CO2. The next day, medium was replaced with 500 µl of the complete growth medium containing [d-Ala2, N-Me-Phe4, Gly5-ol]-enkephalin (DAMGO) or vehicle at a final concentration of 10 µM and incubated for 24 hours at 37°C with 5% CO2. In one set of experiments, cells were pretreated with naltrexone (NTX) (Sigma-Aldrich) or vehicle at a final concentration of 1 µM for 1 hour followed by cotreatment with DAMGO and NTX for 24 hours. After ligand treatment, N2AMOPr cells were washed once with cold phosphate-buffered saline and then collected with 300 µl of cold RLT buffer (QIAGEN, Hilden, Germany). The cell lysates were transferred into an RNase-free centrifuge tube and stored at −80°C until they were further processed for reverse-transcription quantitative polymerase chain reaction (RT-qPCR). Stock solutions of DAMGO (Sigma-Aldrich) and NTX were made in water.
RT-qPCR.
RT-qPCR was performed with the aforementioned N2AMOPr cell lysates and with lysates from mouse brain. In the latter case, we extracted RNA-containing aqueous solution using the TRIzol reagent according to the manufacturer’s protocol (Thermo Fisher Scientific) before the total RNA purification. Total RNA was purified using the RNeasy Mini kit (QIAGEN, Germantown, MD) according to the manufacturer’s protocol. cDNA was synthesized using the PrimeScript RT Master Mix (Takara, Shiga, Japan) according to the manufacturer’s protocol. Real-time PCR was performed using the Power SYBR Green qPCR Master Mix (Applied Biosystems, Foster City, CA). The PCR template source was 4 µl of 10-times diluted first-strand cDNA. Amplification was performed with an ABI PRISM 7900HT sequence detection system (Applied Biosystems) or StepOne Real-Time PCR system (Applied Biosystems). After an initial denaturation step at 95°C for 10 minutes, amplification was performed using 45 cycles of denaturation (95°C for 15 seconds), annealing (55°C for 30 seconds), and extension (72°C for 30 seconds). We amplified glyceraldehyde-3-phosphate dehydrogenase (GAPDH), a housekeeping gene, as a control. The data were analyzed using the sequence detection system software (version 2.2.1, for ABI PRISM 7900HT software, or version 2.3 for StepOne software; Applied Biosystems) as described in Data Analysis. The software generates the baseline-subtracted amplification plot of normalized reporter values (ΔRn) versus cycle number. The amplification threshold was set at six to seven of the ΔRn linear dynamic range (50%–60% of maximum ΔRn). The fractional cycle at which the intersection of the amplification threshold and the plot occurs is defined as the threshold cycle (Ct value) for the plot. Samples that gave a Ct value within 45 cycles were considered to be positive for the mRNA expression. Then, quantitative analysis was performed using the ΔΔCT method, as described previously (Margolis et al., 2014).
The forward (F) and reverse (R) primers were as follows:
GAPDH-F TGAAGGTCGGTGTGAACG
GAPDH-R CAATCTCCACTTTGCCACTG
RTP1-F TGGAAGCCCAGTGAGAAGC
RTP1-R AGCAGAAGTTGCAGCCTGAG
RTP2-F AGCTTTCTGTTCTTCCTTGGG
RTP2-R GCCACCTCCATCTTCTCGTAG
RTP3-F TGCAAGAGGTGAAACCCTGG
RTP3-R AGGACAGTGGAACCTAGCAAAG
RTP4-F GGAGCCTGCATTTGGATAAG
RTP4-R GCAGCATCTGGAACACTGG.
Immunocytochemistry.
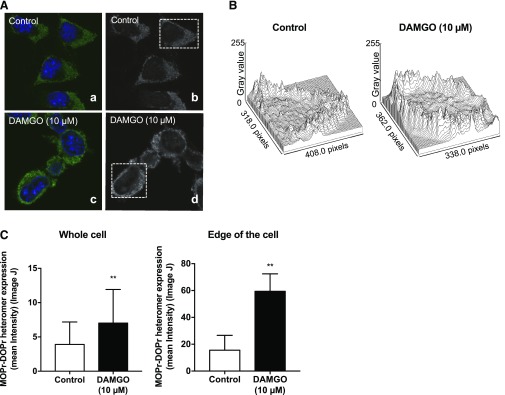
Twenty-four hours after plasmid and/or siRNA transfection, N2AMOPr cells were seeded into a LabTek chamber (20,000 cells/well) in complete growth medium and incubated overnight at 37°C with 5% CO2. The next day, the medium was replaced with 250 µl of the complete growth medium containing DAMGO or vehicle at a final concentration of 10 µM and incubated for 24 hours at 37°C in 5% CO2. After ligand treatment, N2AMOPr cells were fixed with methanol/acetone (1:1) solution for 5 minutes at −30°C and then incubated with blocking buffer containing 3% bovine serum albumin in Tris-buffered saline at room temperature for 1 hour. The cells were then incubated overnight with primary antibody in blocking buffer at 4°C. Mouse anti-MOPr-DOPr heteromer-selective antibody (1:100) (Gupta et al., 2010) or rabbit anti-RTP4 antibody (1:200; MyBioSource, San Diego, CA) was used as the primary antibody. After washing with blocking buffer, three times each for 5 minutes, the cells were incubated with secondary antibody in blocking buffer at room temperature for 1 hour. Goat anti-mouse IgG (H+L) Cross-Adsorbed Alexa Fluor 488 (1:500; Thermo Fisher Scientific) and Goat anti-rabbit IgG (H+L) Cross-Adsorbed Alexa Fluor 594 (1:500; Thermo Fisher Scientific) were used as secondary antibodies. After washing with phosphate-buffered saline, three times each for 5 minutes, cells were mounted with ProLong Diamond Antifade Mounting Medium with 4′,6-diamidino-2-phenylindole (DAPI) (Thermo Fisher Scientific). The immunofluorescent signal was observed under confocal microscopy (LSM800 with Airyscan; Carl Zeiss Meditec, Tokyo, Japan). Surface plots were drawn using ImageJ software (National Institutes of Health, Bethesda, MD). The signal intensity was calculated using the ImageJ software, and the colocalized area was calculated using the Zeiss Zen microscope software (ZEN 2.3 SP1; Carl Zeiss Meditec). When we determined the signal intensity for the whole cell (Fig. 4C), we first outlined the cells and measured the mean intensity of the area using the ImageJ software. On the other hand, when we analyzed the signal intensity on the area close to the edge of the cell (Fig. 4C), the mean signal intensity of 10–12 randomly selected points only on the outline of the cell was measured using the ImageJ software.
Fig. 4.
MOPr-mediated upregulation of RTP4 mRNA leads to increases in MOPr-DOPr heteromer abundance. (A) N2AMOPr cells were treated without or with DAMGO (10 µM) for 24 hour, and the presence of MOPr-DOPr heteromers was detected by immunofluorescence with MOPr-DOPr heteromer-selective antibody as described in Materials and Methods. Images were taken using a Carl Zeiss LSM800 microscope. (B) The surface plot of the cell in the dotted square in (A) was analyzed using ImageJ software. (C) The signal intensity of the MOPr-DOPr heteromer was measured by ImageJ software. n = 196 (control), n = 341 (DAMGO 10 µM) (whole cell), n = 205 (control), n = 381 (DAMGO 10 µM) (the area close to the edge of the cell). **P < 0.01 vs. control, unpaired t test.
Animals.
Male C57BL/6 mice (25–35 g; 6–12 weeks) were obtained from Jackson Laboratories (Bar Harbor, ME). All mice were maintained on a 12-hour light/dark cycle with rodent chow and water available ad libitum, and they were housed in groups of five until testing. Animal studies were carried out according to protocols approved by the Icahn School of Medicine at Mount Sinai Animal Care and Use Committee.
Drug Administration.
Morphine sulfate (Sigma-Aldrich) was dissolved in saline. Male C57BL/6 mice were injected with morphine sulfate (10 mg/kg) subcutaneously once a day for 10 days. On the 11th day, mice were euthanized by cervical dislocation, and various brain regions were collected and transferred into an RNase-free centrifuge tube and stored at −80°C until they were further processed for RT-qPCR.
Enzyme-Linked Immunosorbent Assay.
Enzyme-linked immunosorbent assay (ELISA) was carried out as previously described (Gupta et al., 2010; Fujita et al., 2014). In brief, mouse brain regions were dissected according to the mouse brain atlas (Paxinos and Franklin, 2001), and the membrane fraction (10 µg) was subjected to ELISA using either mouse anti-MOPr-DOPr heteromer-selective antibody (1:100), rat anti-MOPr antibody (1:500), rat anti-DOPr antibody (1:500), rat anti-CB1 receptor antibody (1:500), or rabbit anti-D2 receptor antibody (1:500) as primary antibodies. As secondary antibodies, anti-mouse IgG (1:1000; Vector Laboratories, Burlingame, CA), anti-rat IgG (1:1000; Jackson ImmunoResearch Laboratories, West Grove, PA), or anti-rabbit IgG (1:1000; Vector Laboratories) coupled to horseradish peroxidase was used as described previously (Gupta et al., 2007). ELISA for each sample was performed in triplicate.
Data Analysis.
The data were expressed as the mean ± S.D. Student’s t test, one-way analysis of variance (ANOVA), two-way ANOVA, one-way multivariate ANOVA, two-way multivariate ANOVA, and multiple-comparison test (Newman-Keuls test, Tukey’s test, or Bonferroni test) were used to analyze the data. A difference was considered to be significant at P < 0.05. GraphPad Software Prism7 (GraphPad Software, La Jolla, CA) and JMP Software 14 (SAS Institute Inc., Cary, NC) were used.
Results
In this study, we investigated the effect of chronic morphine administration in mice on the levels of RTP mRNAs in different regions of the brain. In addition, we examined the role of RTP4 in regulation of MOPr-DOPr heteromer formation following MOPr activation in vivo and in vitro.
RTP4 Is Highly Expressed in Mouse Brain.
We first surveyed the gene-expression data sets of the mouse brain (based on Affymetrix experiments that can be accessed using BioGPS, http://biogps.org) for relative levels of expression of RTPs. This analysis showed that RTPs (RTP1–4) are differentially expressed in various brain regions, and that among the various RTPs, RTP4 exhibited the highest expression (Supplemental Fig. 1, upper panel). Next, we quantified the levels of RTP4 in different brain regions by quantitative RT-qPCR. Data from this analysis revealed over 2-fold higher expression of the RTP4 mRNA in midbrain, nucleus accumbens (NAcc), pons, hypothalamus, and ventral tegmental area (VTA) compared with the cortex; other brain regions had lower levels (Supplemental Fig. 1, lower panel).
RTP4 Expression Is Regulated by Chronic Morphine Administration and This Affects Receptor Abundance of MOPr and MOPr-DOPr Heteromer in Select Brain Regions.
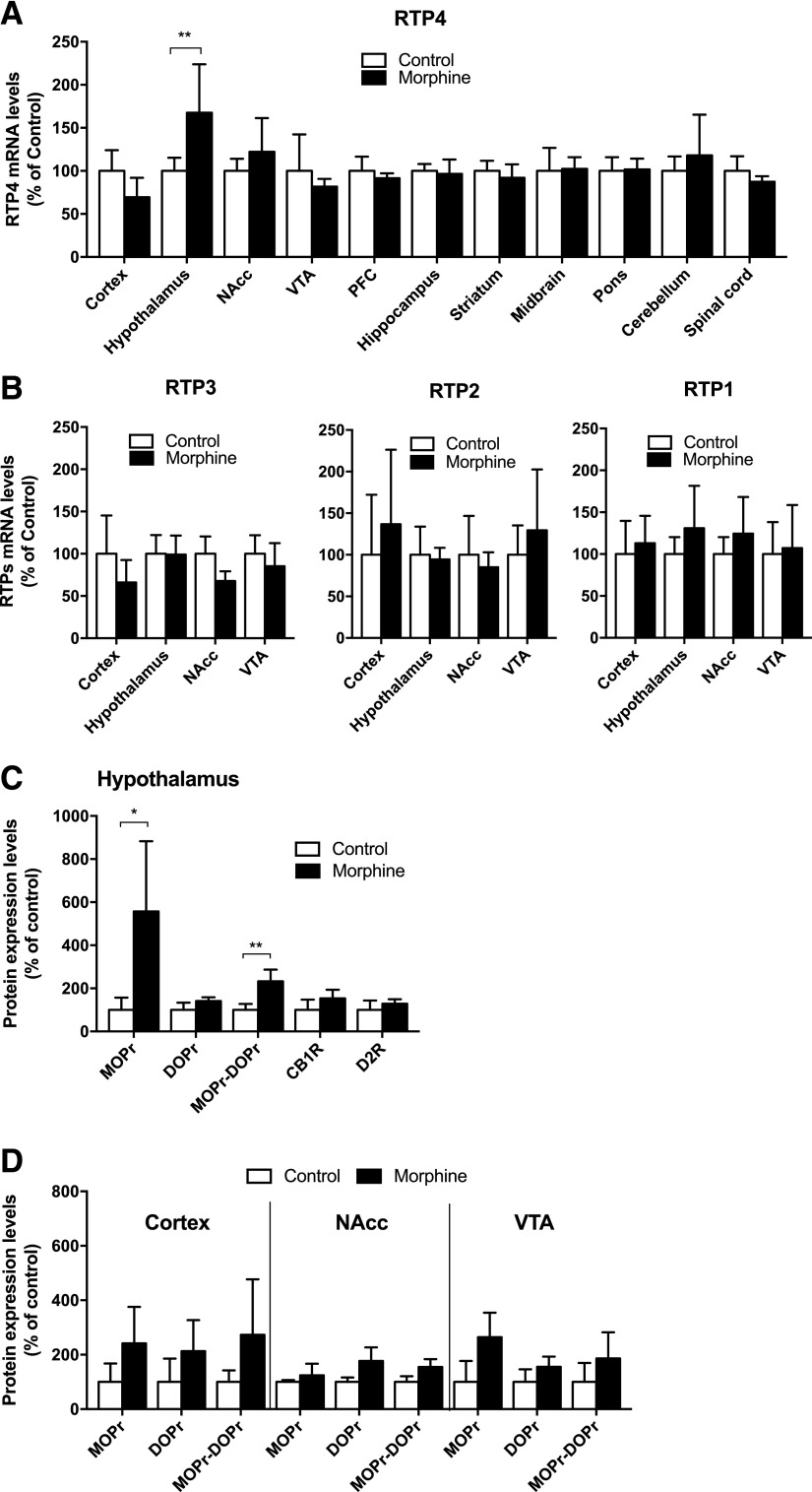
Next, we examined whether administration of morphine to mice affected RTP4 mRNA levels in the brain. For this, mice were subjected to repeated morphine administration (10 mg/kg, s.c., once a day for 10 days), after which RTP4 mRNA levels in various brain regions were quantified by RT-qPCR. We detected a significant increase (P < 0.05) of RTP4 mRNA levels only in the hypothalamus among the different brain regions tested (Fig. 1A). There were no significant changes in the mRNA levels of RTP1–3 except for a small decrease in RTP3 mRNA levels in the nucleus accumbens of morphine-treated animals (Fig. 1B). Next, we examined whether the increases in RTP4 mRNA levels led to an increase in membrane levels of MOPr-DOPr heteromers in the hypothalamus. For this, we carried out ELISAs with antibodies that selectively recognize receptors in endogenous tissue and compared the abundance of these receptors in hypothalamic membranes from saline-treated animals to the receptors in the hypothalamic membranes of morphine-treated animals (10 mg/kg, s.c., once a day for 10 days); as controls, we used membranes from the cortex, nucleus accumbens, and VTA. Previous studies characterizing these antibodies used tissues from animals lacking individual receptors to demonstrate the specificity of these antibodies (Gupta et al., 2010). In this study, we found that repeated morphine administration leads to a significant increase (P < 0.05) in MOPr-DOPr heteromer and MOPr levels but not DOPr levels or levels of CB1 cannabinoid receptors or D2 dopamine receptors (Fig. 1C). This increase was only seen in the hypothalamus and not in the cortex, NAcc, and VTA (Fig. 1D).
Fig. 1.
RTP4 expression is regulated by chronic morphine administration, and this affects abundance of MOPr and MOPr-DOPr heteromers. (A and B) Male C57BL/6 mice (5–6/group) were administered saline or morphine (10 mg/kg, s.c.) once a day for 10 days. On the 11th day, brain regions were collected and RT-qPCR was performed. Data were normalized with GAPDH, an internal control. Data are the mean ± S.D. n = 5–6. **P < 0.01 vs. control group (saline), two-way ANOVA and Bonferroni test (A) or two-way multivariate ANOVA (B). (C and D) Male C57BL/6 mice (5–6/group) were administered saline or morphine (10 mg/kg, s.c.) once a day for 10 days. On the 11th day, the hypothalamus, cortex, NAcc, and VTA were collected. Membrane protein (10 µg) was subjected to ELISA using receptor-specific antibodies. Data are the mean ± S.D. n = 3–6. *P < 0.05; **P < 0.01 vs. control group (saline), unpaired t test with Welch’s correction (C) or two-way multivariate ANOVA (D). PFC = prefrontal cortex.
MOPr Activation in N2AMOPr Cells Causes Significant Upregulation of RTP4 mRNA Levels.
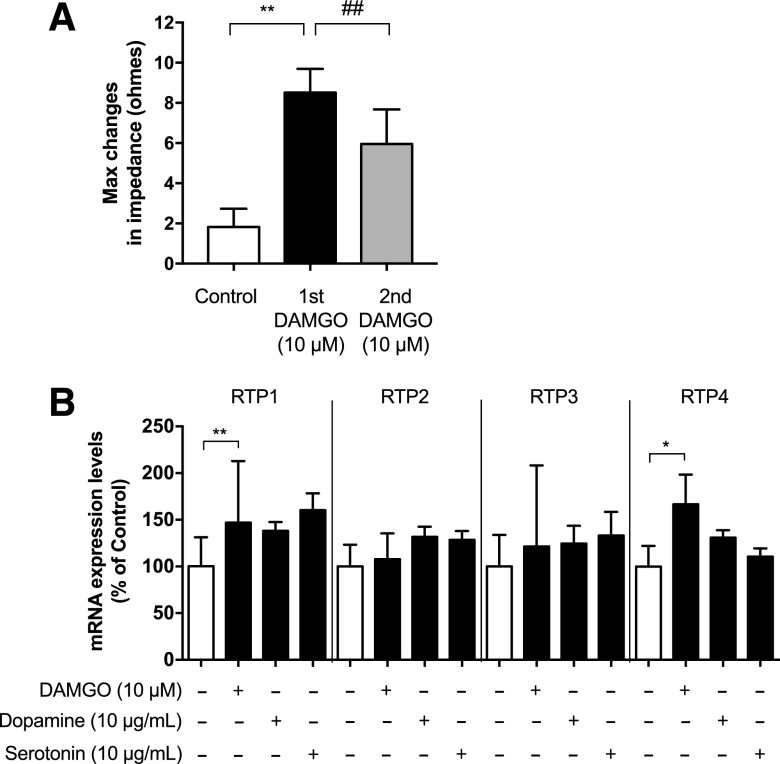
Next, we used Neuro 2A cells transfected with MOPr (N2AMOPr cells) as a model system to explore the molecular mechanisms underlying the upregulation of RTP4 mRNA levels to determine if the RTP4 effects seen in vivo are a circuit-based or a cell-based phenomenon. In N2AMOPr cells, we confirmed that DAMGO, the MOPr-selective agonist, activates Gαi-mediated signaling using the CellKey System, which measures complex impedance changes in cell monolayers after GPCR activation (increase in impedance means Gαi activation) (Fig. 2A) and found that this can be blocked by the selective MOPr antagonist, NTX (Fig. 3A). A second pulse of DAMGO 24 hours after first exposure to the drug gave a significantly reduced (P < 0.01) Gαi activation (Fig. 2A, second DAMGO), suggesting the development of tolerance to the DAMGO-induced Gαi response.
Fig. 2.
MOPr activation in N2AMOPr cells causes significant upregulation of RTP4 mRNA levels. (A) N2AMOPr cells (3.2 × 104 cells/well) were treated without or with DAMGO (10 µM) for 20 minutes (first DAMGO) and the Gαi response was measured using the CellKey System. After 24 hours, cells were given a second treatment with DAMGO (10 µM) (second DAMGO) followed by measurement of the Gαi response. Data are the mean ± S.D. n = 8–9. **P < 0.01 vs. control; ##P < 0.01 vs. first DAMGO, Tukey’s test. (B) N2AMOPr cells (2 × 104 cells/well) were treated with DAMGO (10 µM), dopamine (10 µg/ml), or serotonin (10 µg/ml) for 24 hours. RT-qPCR was performed with specific primers against RTP1, 2, 3, 4, and GAPDH. Data are the mean ± S.D. n = 7–45. **P < 0.01; *P < 0.05 vs. control, one-way multivariate ANOVA and Bonferroni test.
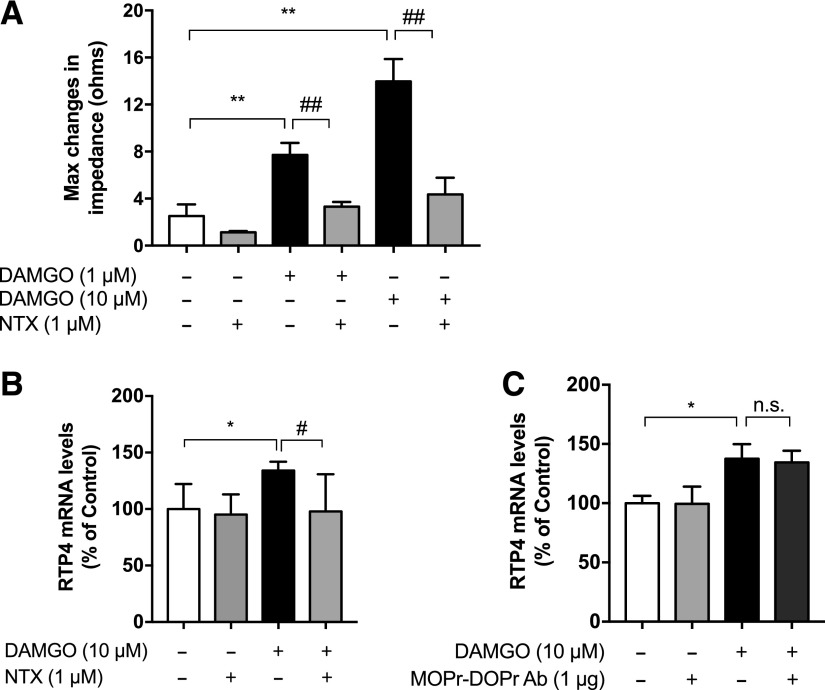
Fig. 3.
Activation of MOPr in N2AMOPr cells upregulates RTP4 mRNA levels. (A) N2AMOPr cells (3.2 × 104 cells/well) were treated with DAMGO (1 or 10 µM) with or without pretreatment with NTX (1 µM) as described in Materials and Methods, and the Gαi response was measured using the CellKey System. Data are the mean ± S.D. n = 3–12. **P < 0.01 vs. control; ##P < 0.01 vs. DAMGO alone, Tukey’s test. (B) N2AMOPr cells (2 × 104 cells/well) were treated without or with DAMGO (10 µM) for 24 hours with or without pretreatment with NTX (1 µM) for 1 hour before addition of DAMGO. RT-qPCR was performed with specific primers against RTP4 and GAPDH. Data are the mean ± S.D. n = 6–11. *P < 0.05 vs. control; #P < 0.05 vs. DAMGO alone, Tukey’s test. (C) N2AMOPr cells (2 × 104 cells/well) were treated with DAMGO (10 µM) for 24 hours without or with pretreatment with MOPr-DOPr heteromer-selective antibody (Ab; 1 µg) for 1 hour before addition of DAMGO. RT-qPCR was performed with specific primers against RTP4 and GAPDH. Data are the mean ± S.D. n = 6–15; *P < 0.05 vs. control, Tukey’s test. n.s., not significant.
Next, we examined if, similar to morphine, DAMGO could induce changes in the gene expression of RTP4. For this, we treated N2AMOPr cells with DAMGO for 24 hours and found that this led to a significant increase (P < 0.05) in RTP4 mRNA levels (Fig. 2B). We also examined whether DAMGO treatment modulated the levels of other RTPs (RTP1–3) and found that it significantly increased (P < 0.01) the mRNA levels of RTP1 but not RTP2 or RTP3 (Fig. 2B). Interestingly, activation of other class A GPCRs known to be expressed in Neuro 2A cells (http://biogps.org) (e.g., dopamine and serotonin receptors) by their respective agonists did not cause significant increases in either RTP4 or the related RTPs (RTP 1–3; Fig. 2B). In addition, we found that DAMGO-mediated increases in RTP4 mRNA levels can be blocked by pretreating N2AMOPr cells with NTX (Fig. 3B). Next, we examined if pretreatment with the MOPr-DOPr heteromer-selective antibody affected DAMGO-mediated increases in RTP4 mRNA levels and found that this pretreatment had no significant effect on RTP4 mRNA levels (Fig. 3C). Since the MOPr-DOPr heteromer-selective antibody has been shown to selectively block heteromer-mediated signaling (Gupta et al., 2010), these results suggest that DAMGO-mediated increases in RTP4 mRNA levels occur via MOPr.
MOPr-Mediated Upregulation of RTP4 mRNA Levels Leads to Increases in MOPr-DOPr Abundance.
Next, we examined if DAMGO-mediated increases in RTP4 mRNA levels lead to increases in the abundance of the MOPr-DOPr heteromer. To detect cell surface levels, we carried out immunofluorescence analysis using the MOPr-DOPr heteromer-selective antibody in N2AMOPr cells treated without or with DAMGO (10 μM) for 24 hours. This analysis revealed that the MOPr-DOPr heteromer exhibits a broad punctate distribution within the cells (Fig. 4A). The surface plot analyzed by the ImageJ software using the black/white images (Fig. 4A) revealed that the signal intensity of the MOPr-DOPr heteromer in DAMGO-treated cells was greater than that of control cells (Fig. 4B). The quantitative analysis showed a significant increase (P < 0.01) in the signal intensity of the MOPr-DOPr heteromer in the entire area of the cell 24 hours after treatment with DAMGO (Fig. 4C, whole cell). In addition, the signal intensity of the MOPr-DOPr heteromer in the selected area close to the edge of the cell was also significantly increased (Fig. 4C, edge of the cell). These results indicate that prolonged treatment with DAMGO leads to increased MOPr-DOPr heteromer abundance.
MOPr-Mediated Increases in MOPr-DOPr Heteromer Abundance Are Dependent on RTP4.
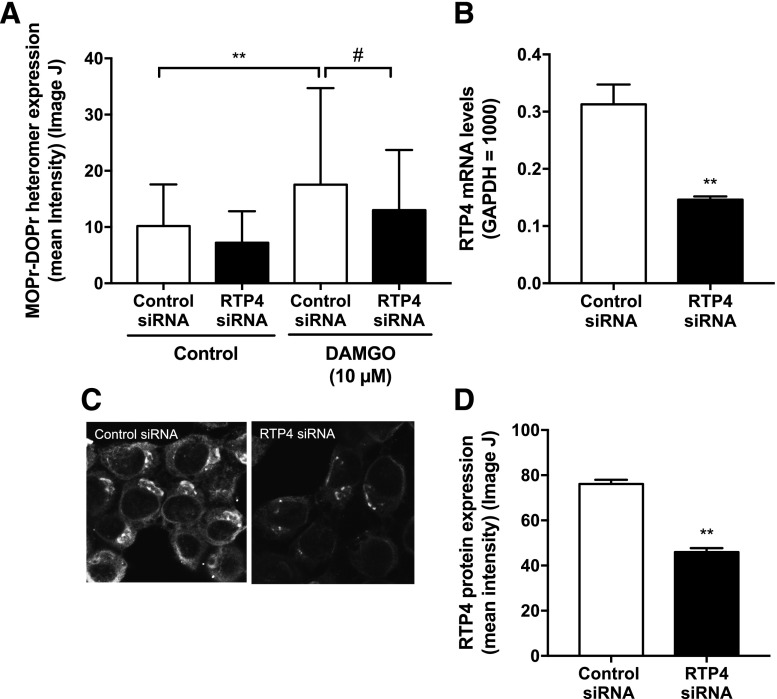
Given that DAMGO treatment leads to increases in RTP4 mRNA levels as well as MOPr-DOPr heteromer abundance, we examined if RTP4 is required for heteromer upregulation. For this, we pretreated N2AMOPr cells with RTP4 siRNA prior to DAMGO treatment. We found that this treatment led to no significant increase in the abundance of MOPr-DOPr heteromer in response to DAMGO treatment; in contrast, an increase was seen in cells treated with the control siRNA (Fig. 5A). We confirmed that RTP4 siRNA treatment led to a significant decrease in the levels of RTP4 mRNA (Fig. 5B) and protein (Fig. 5, C and D). These results, showing that a decrease in the expression of RTP4 leads to decreases in MOPr-DOPr levels, indicate that RTP4 plays a role in the maturation of MOPr-DOPr heteromers.
Fig. 5.
MOPr-mediated increases in MOPr-DOPr heteromer abundance are dependent on RTP4 expression. (A) N2AMOPr cells were transfected with either RTP4 siRNA or control siRNA and then treated with or without DAMGO (10 µM) for 24 hours. Expression levels of MOPr-DOPr heteromers in the area close to the edge of the cell were analyzed by immunofluorescence using MOPr-DOPr heteromer-selective antibodies. Images were taken using a Carl Zeiss LSM800 microscope. n = 47 (control siRNA), n = 20 (RTP4 siRNA), n = 47 (control siRNA + 10 µM DAMGO), n = 67 (RTP4 siRNA + 10 µM DAMGO). **P < 0.01 vs. control-control siRNA; #P < 0.05 vs. DAMGO-control siRNA, Newman-Keuls test. (B–D) RTP4 siRNA expression decreases RTP4 mRNA and protein levels. N2AMOPr cells were transfected with RTP4 siRNA or control siRNA. After 48 hours, RTP4 mRNA levels (B) were analyzed by RT-qPCR (n = 3; **P < 0.01 vs. control siRNA, unpaired t test) and RTP4 protein expression levels (C and D) by immunofluorescence using an RTP4-selective antibody. (C) Images were taken using a Carl Zeiss LSM800 microscope. (D) The signal intensity of RTP4 was measured using ImageJ software. n = 144 (control siRNA), n = 188 (RTP4 siRNA). **P < 0.01 vs. control siRNA, unpaired t test.
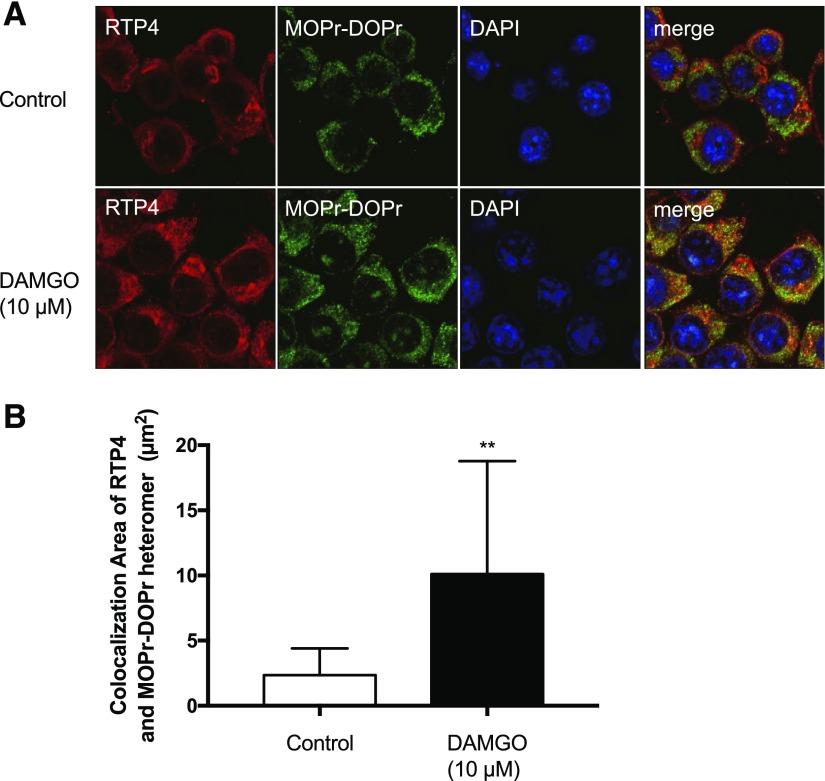
MOPr-Mediated Upregulation of RTP4 Increases Colocalization of MOPr-DOPr Heteromer with RTP4.
We also examined if treatment of N2AMOPr cells with DAMGO for 24 hours increased colocalization between RTP4 and MOPr-DOPr heteromers. For this, we costained the cells with anti-RTP4 and with MOPr-DOPr heteromer-selective antibodies and examined the extent of their colocalization. As expected, in control cells, RTP4 appears to be localized to Golgi- and endoplasmic reticulum-like intracellular compartments (Fig. 6A, upper panel). Interestingly, upon treatment with DAMGO (10 µM for 24 hours), we found that RTP4 was detected in the cytosol (Fig. 6A, lower panel). Moreover, we found a significant increase in the colocalization of MOPr-DOPr heteromer with RTP4 (i.e., the area of merged yellow signal) upon DAMGO treatment (Fig. 6B). These results indicate an increased association between RTP4 and MOPr-DOPr heteromers, which would support a role for RTP4 in the maturation of this heteromer.
Fig. 6.
Increase in colocalization between RTP4 and the MOPr-DOPr heteromer following MOPr activation. N2AMOPr cells were treated without or with DAMGO (10 µM) for 24 hours and analyzed by immunofluorescence using the MOPr-DOPr heteromer-selective antibody and an RTP4-selective antibody. (A) Images were taken using a Carl Zeiss LSM800 microscope (63× oil lens, Airyscan). Red, RTP4; green, MOPr-DOPr heteromer; blue, 4′,6-diamidino-2-phenylindole (DAPI). (B) Colocalized area indicated by yellow signal in the merged image was examined using ZEN software (Carl Zeiss microscope). n = 23 (control), n = 26 (DAMGO). **P < 0.01 vs. control, unpaired t test.
Discussion
In this study, we explored a role for RTP family members, previously shown to be involved in targeting odorant and taste receptors to the cell surface (Saito et al., 2004; Behrens et al., 2006), as a chaperone of opioid receptors. Analysis of the gene expression data sets showed that these proteins are widely expressed in the brain and periphery (http://biogps.org). Among the different RTPs, RTP4 was previously shown to help target MOPr and DOPr to the cell surface in either HEK-293 or Neuro 2A cells (Décaillot et al., 2008). In this study, we found that repeated morphine administration leads to brain-region-specific modulation of RTP4 mRNA levels, with increased levels being observed in the hypothalamus. Other than RTP4, the only other RTPs that were found to be regulated by repeated morphine treatment were RTP3, whose levels showed a trend of decrease in the NAcc and in the cortex; RTP2, which showed a small trend of increase in the cortex and VTA; and RTP1, which showed a small trend of increase in the hypothalamus and NAcc. These results suggest a brain-area-specific modulation of RTP levels by morphine. Although very little is known about how morphine regulates RTP4 levels, one possibility is that repeated administration leads to region-specific adaptations that affect the levels of the transcription factors that bind to the promoter region of RTP4, leading to up-/downregulation of mRNA levels. Support for this comes from studies showing that chronic morphine treatment leads to differential activation of transcription factors in different areas of the brain, and this may be dependent on the schedule and dose of morphine administration [reviewed by Ammon-Treiber and Höllt (2005)]. Another possibility is that the region-specific adaptations induced by repeated morphine administration affect the stability of the mRNA for RTP4. It is also possible that the adaptations to repeated morphine administration could be brought about by brain-region-specific modulation of MOPr signaling involving either β-arrestin or mitogen-activated protein kinase signaling instead of the normal Gαi-mediated signaling (Al-Hasani and Bruchas, 2011; Williams et al., 2013). Thus, further studies are needed to elucidate how morphine regulates RTP4 levels in the brain. In addition, with regard to antinociception, it is well known that the hypothalamus regulates the endogenous opioid ligand-mediated descending inhibitory pathway projecting from periaqueductal gray to the nociceptive neurons located in the dorsal horn of the spinal cord and, thus, controls nociceptive input from the periphery (Basbaum and Fields, 1979). It is possible that after chronic morphine administration, the endogenous compensatory mechanisms against excessive activation of the opioid system might occur in the hypothalamus. Hypothalamus-specific knockdown of RTP4 on the development of antinociceptive tolerance to morphine would have to be evaluated in future studies.
Although we found that repeated morphine administration modulates RTP4 levels in specific brain regions, this does not rule out the modulation of the levels of other chaperone proteins in other brain areas expressing MOPr. This could account for the observation that, although levels of RTP4 were decreased in the cortex, MOPr, DOPr, and MOPr-DOPr heteromers were increased. Candidate chaperone proteins for MOPr could include ribophorin I (Ge et al., 2009), which has been shown to target MOPr, or proteins such as DRiP78, ATBP50, RACK1, GEC1, norbin, MRAP, or USP4, which have been shown to help in the cell surface targeting of other GPCRs [reviewed by Williams and Devi (2010)]. Members of the ubiquitin-specific protease family (USP) would be interesting candidates since USP4 has been shown to function as an adenosine A2A receptor chaperone, helping target it to the cell surface and protect this receptor from ubiquitination and degradation (Milojevic et al., 2006). We previously found that USP8, a member of this family, is downregulated in the quantitative proteomic studies with striatal postsynaptic density fractions of mice treated with escalating doses of morphine for 5 days (Stockton et al., 2015). Thus, further studies are needed to determine the brain-region-specific putative MOPr chaperone proteins that are modulated following chronic treatment with morphine and how the levels of these chaperones are affected under the various paradigms of chronic morphine administration.
Our observations that increased RTP4 mRNA levels led to increases in protein levels of MOPr but not DOPr, CB1 receptor, or D2 receptor in the hypothalamus would suggest that only certain proteins could function as a chaperone for a given GPCR. This is supported by our data in N2AMOPr cells, where we found that activation of dopamine receptors does not lead to significant increases in RTP mRNA levels. Moreover, studies have shown that RACK1 helps target thromboxane A2 and chemokine CXCR4 receptors to the cell surface but not β2-adrenergic or prostanoid prostaglandin (PG) D2 receptor (DP) receptors (Parent et al., 2008), and GEC1 facilitates the trafficking of Kappa opioid receptor (KOPr), but not MOPr or DOPr, to the cell surface (Chen et al., 2006). The robust increases in levels of MOPr protein in the hypothalamus following chronic morphine treatment could be due to an increase in its mRNA levels. However, qRT-PCR analysis did not detect significant changes in MOPr mRNA levels (control = 100% ± 36; morphine treated = 96% ± 50; data not shown). This would indicate that RTP4 could play a stabilizing effect on MOPr protein level and prevent it from degradation.
In this study, we found that repeated morphine administration, in addition to modulating RTP4 mRNA levels, significantly increases expression of MOPr-DOPr heteromers only in the hypothalamus. This is in contrast to a previous study showing that chronic morphine administration leads to significant increases in MOPr-DOPr heteromers in brain regions involved in pain processing and in the spinal cord (Gupta et al., 2010). These differences could be due to the different paradigms of morphine administration or additional mechanisms regulating MOPr-DOPr heteromer levels. Our in vitro data indicate that RTP4 plays a role in increased MOPr-DOPr heteromer expression. In addition, the control experiments suggest that the maturation of the MOPr-DOPr heteromer under physiologic conditions requires endogenous RTP4. However, this does not rule out the possibility that there might be both RTP4-dependent and RTP4-independent mechanisms in the increased MOPr-DOPr heteromer formation in vivo. In addition, the in vitro study revealed the possibility of the involvement of RTP4 in the mechanism of development of tolerance to the DAMGO-induced Gαi response, which may relate to the side effects of chronic morphine treatment, such as development of analgesic tolerance in vivo. Thus, further studies are needed to explore mechanisms regulating MOPr-DOPr heteromer expression in different brain regions. In addition, we found that the protein level of MOPr showed a robust (i.e., >5-fold) increase, whereas the increase of MOPr-DOPr was around 2-fold, and there were no changes in DOPr protein levels in the hypothalamus after chronic morphine treatment (Fig. 1C). These results suggest the possibility that the increase in MOPr-DOPr heteromer formation would depend on the increase of cell surface MOPr. However, as is well known, prior works on the effects of chronic morphine treatment on regulation of MOPr abundance are controversial. Some papers have suggested an increase of plasma membrane binding of MOPr agonist (Fábián et al., 2002), whereas others have reported downregulation of MOPr mRNA levels in the mouse brain (Zhu et al., 2012) or no change in MOPr protein levels (Ujcikova et al., 2017). It is probable that these differences are due to the experimental paradigm or brain regions used, as a recent article suggested that the changes in mRNA levels depend on the MOPr splicing variants and on the brain regions (Xu et al., 2015) used.
In addition to serving as a chaperone protein for the transport of GPCRs (Behrens et al., 2006; Décaillot et al., 2008; Gifford et al., 2008), RTP4 has been shown to have other physiologic roles. RTP4 is one of the interferon-stimulated genes increased in peripheral blood leukocytes after pregnancy and, thus, could be used as a marker of pregnancy, particularly in rodents (Gifford et al., 2008; Ribeiro et al., 2014; Kose et al., 2016; Sinedino et al., 2017). In addition, RTP4 plays an important role as a marker of disease incidence in conditions such as hypertension in type 2 diabetic patients, Q-fever, interferon-induced depression, HER2(+) breast cancer, and virus infection (Laurin et al., 2013; Mehraj et al., 2013; Davenport et al., 2015; Hoyo-Becerra et al., 2015; Radkowski et al., 2016). Studies have also indicated that in addition to having antiviral activity, RTP4 is involved in microbiota regulation and inflammatory cytokine secretion (Schoggins et al., 2011; Ueta et al., 2011; Brodziak et al., 2013). It is not clear if RTP4 functions as a chaperone under these various scenarios. All of these roles underscore the necessity to understand how RTP4 levels are regulated under different conditions and suggest that it could be a target for the development of therapeutics for disease conditions in which the levels of this protein are altered.
In conclusion, in this study, we found that prolonged MOPr activation leads to increases in RTP4 mRNA levels in mouse brain and N2AMOPr cells. Moreover, we found that in N2AMOPr cells, RTP4 is necessary and sufficient to increase cell surface expression of MOPr-DOPr heteromers. This suggests that RTP4 could be a putative therapeutic target for treatment of pain or side effects involving MOPr and/or MOPr-DOPr heteromers.
Acknowledgments
We thank Nikita Trimbake for help with the RT-qPCR assay.
Abbreviations
- ANOVA
analysis of variance
- DAMGO
[d-Ala2, N-Me-Phe4, Gly5-ol]-enkephalin
- DOPr
delta opioid receptor
- ELISA
enzyme-linked immunosorbent assay
- F
forward
- GAPDH
glyceraldehyde-3-phosphate dehydrogenase
- GPCR
G protein–coupled receptor
- MOPr
mu opioid receptor
- NAcc
nucleus accumbens
- NTX
naltrexone
- R
reverse
- RTP
receptor transporter protein
- RT-qPCR
reverse-transcription quantitative polymerase chain reaction
- siRNA
small interfering RNA
- USP
ubiquitin-specific protease family
- VTA
ventral tegmental area
Authorship Contributions
Participated in research design: Fujita, Devi.
Conducted experiments: Fujita, Yokote, Gupta.
Performed data analysis: Fujita, Yokote, Gomes.
Wrote or contributed to the writing of the manuscript: Fujita, Gomes, Ueda, Devi.
Footnotes
This work was supported by the Japan Society for the Promotion of Science KAKENHI [Grants 16K19214 and 18K06894]; the Platform for Drug Discovery, Informatics, and Structural Life Science [Grant 16am0101012j0005] from the Japan Agency for Medical Research and Development (AMED); and the National Institutes of Health [Grant DA008863].
 This article has supplemental material available at molpharm.aspetjournals.org.
This article has supplemental material available at molpharm.aspetjournals.org.
References
- Achour L, Labbé-Jullié C, Scott MG, Marullo S. (2008) An escort for GPCRs: implications for regulation of receptor density at the cell surface. Trends Pharmacol Sci 29:528–535. [DOI] [PubMed] [Google Scholar]
- Al-Hasani R, Bruchas MR. (2011) Molecular mechanisms of opioid receptor-dependent signaling and behavior. Anesthesiology 115:1363–1381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammon-Treiber S, Höllt V. (2005) Morphine-induced changes of gene expression in the brain. Addict Biol 10:81–89. [DOI] [PubMed] [Google Scholar]
- Basbaum AI, Fields HL. (1979) The origin of descending pathways in the dorsolateral funiculus of the spinal cord of the cat and rat: further studies on the anatomy of pain modulation. J Comp Neurol 187:513–531. [DOI] [PubMed] [Google Scholar]
- Behrens M, Bartelt J, Reichling C, Winnig M, Kuhn C, Meyerhof W. (2006) Members of RTP and REEP gene families influence functional bitter taste receptor expression. J Biol Chem 281:20650–20659. [DOI] [PubMed] [Google Scholar]
- Brodziak F, Meharg C, Blaut M, Loh G. (2013) Differences in mucosal gene expression in the colon of two inbred mouse strains after colonization with commensal gut bacteria. PLoS One 8:e72317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Li JG, Chen Y, Huang P, Wang Y, Liu-Chen LY. (2006) GEC1 interacts with the kappa opioid receptor and enhances expression of the receptor. J Biol Chem 281:7983–7993. [DOI] [PubMed] [Google Scholar]
- Davenport EE, Antrobus RD, Lillie PJ, Gilbert S, Knight JC. (2015) Transcriptomic profiling facilitates classification of response to influenza challenge. J Mol Med (Berl) 93:105–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Décaillot FM, Rozenfeld R, Gupta A, Devi LA. (2008) Cell surface targeting of mu-delta opioid receptor heterodimers by RTP4. Proc Natl Acad Sci USA 105:16045–16050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fábián G, Bozó B, Szikszay M, Horváth G, Coscia CJ, Szücs M. (2002) Chronic morphine-induced changes in mu-opioid receptors and G proteins of different subcellular loci in rat brain. J Pharmacol Exp Ther 302:774–780. [DOI] [PubMed] [Google Scholar]
- Fujita W, Gomes I, Dove LS, Prohaska D, McIntyre G, Devi LA. (2014) Molecular characterization of eluxadoline as a potential ligand targeting mu-delta opioid receptor heteromers. Biochem Pharmacol 92:448–456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ge X, Loh HH, Law PY. (2009) mu-Opioid receptor cell surface expression is regulated by its direct interaction with ribophorin I. Mol Pharmacol 75:1307–1316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gifford CA, Assiri AM, Satterfield MC, Spencer TE, Ott TL. (2008) Receptor transporter protein 4 (RTP4) in endometrium, ovary, and peripheral blood leukocytes of pregnant and cyclic ewes. Biol Reprod 79:518–524. [DOI] [PubMed] [Google Scholar]
- Gomes I, Ijzerman AP, Ye K, Maillet EL, Devi LA. (2011) G protein-coupled receptor heteromerization: a role in allosteric modulation of ligand binding. Mol Pharmacol 79:1044–1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes I, Jordan BA, Gupta A, Trapaidze N, Nagy V, Devi LA. (2000) Heterodimerization of mu and delta opioid receptors: a role in opiate synergy. J Neurosci 20:RC110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Décaillot FM, Gomes I, Tkalych O, Heimann AS, Ferro ES, Devi LA. (2007) Conformation state-sensitive antibodies to G-protein-coupled receptors. J Biol Chem 282:5116–5124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Mulder J, Gomes I, Rozenfeld R, Bushlin I, Ong E, Lim M, Maillet E, Junek M, Cahill CM, et al. (2010) Increased abundance of opioid receptor heteromers after chronic morphine administration. Sci Signal 3:ra54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoyo-Becerra C, Liu Z, Yao J, Kaltwasser B, Gerken G, Hermann DM, Schlaak JF. (2015) Rapid regulation of depression-associated genes in a new mouse model mimicking interferon-α-related depression in hepatitis C virus infection. Mol Neurobiol 52:318–329. [DOI] [PubMed] [Google Scholar]
- Katada S, Tanaka M, Touhara K. (2004) Structural determinants for membrane trafficking and G protein selectivity of a mouse olfactory receptor. J Neurochem 90:1453–1463. [DOI] [PubMed] [Google Scholar]
- Kose M, Kaya MS, Aydilek N, Kucukaslan I, Bayril T, Bademkiran S, Kiyma Z, Ozyurtlu N, Kayis SA, Guzeloglu A, et al. (2016) Expression profile of interferon tau-stimulated genes in ovine peripheral blood leukocytes during embryonic death. Theriogenology 85:1161–1166. [DOI] [PubMed] [Google Scholar]
- Laurin M, Huber J, Pelletier A, Houalla T, Park M, Fukui Y, Haibe-Kains B, Muller WJ, Côté JF. (2013) Rac-specific guanine nucleotide exchange factor DOCK1 is a critical regulator of HER2-mediated breast cancer metastasis. Proc Natl Acad Sci USA 110:7434–7439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margolis EB, Hjelmstad GO, Fujita W, Fields HL. (2014) Direct bidirectional μ-opioid control of midbrain dopamine neurons. J Neurosci 34:14707–14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsunami H, Mainland JD, Dey S. (2009) Trafficking of mammalian chemosensory receptors by receptor-transporting proteins. Ann N Y Acad Sci 1170:153–156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehraj V, Textoris J, Ben Amara A, Ghigo E, Raoult D, Capo C, Mege JL. (2013) Monocyte responses in the context of Q fever: from a static polarized model to a kinetic model of activation. J Infect Dis 208:942–951. [DOI] [PubMed] [Google Scholar]
- Milojevic T, Reiterer V, Stefan E, Korkhov VM, Dorostkar MM, Ducza E, Ogris E, Boehm S, Freissmuth M, Nanoff C. (2006) The ubiquitin-specific protease Usp4 regulates the cell surface level of the A2A receptor. Mol Pharmacol 69:1083–1094. [DOI] [PubMed] [Google Scholar]
- Parent A, Laroche G, Hamelin E, Parent JL. (2008) RACK1 regulates the cell surface expression of the G protein-coupled receptor for thromboxane A(2). Traffic 9:394–407. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Franklin KBJ. (2001) The Mouse Brain in Stereotaxic Coordinates, 2nd ed, Academic Press, Cambridge, MA. [Google Scholar]
- Radkowski P, Wątor G, Skupien J, Bogdali A, Wołkow P. (2016) Analysis of gene expression to predict dynamics of future hypertension incidence in type 2 diabetic patients. BMC Proc 10 (Suppl 7):113–117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro ES, Bruno RG, Farias AM, Hernández-Rivera JA, Gomes GC, Surjus R, Becker LF, Birt A, Ott TL, Branen JR, et al. (2014) Low doses of bovine somatotropin enhance conceptus development and fertility in lactating dairy cows. Biol Reprod 90:10. [DOI] [PubMed] [Google Scholar]
- Rozenfeld R, Devi LA. (2007) Receptor heterodimerization leads to a switch in signaling: beta-arrestin2-mediated ERK activation by mu-delta opioid receptor heterodimers. FASEB J 21:2455–2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saito H, Kubota M, Roberts RW, Chi Q, Matsunami H. (2004) RTP family members induce functional expression of mammalian odorant receptors. Cell 119:679–691. [DOI] [PubMed] [Google Scholar]
- Schoggins JW, Wilson SJ, Panis M, Murphy MY, Jones CT, Bieniasz P, Rice CM. (2011) A diverse range of gene products are effectors of the type I interferon antiviral response. Nature 472:481–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinedino LD, Honda PM, Souza LR, Lock AL, Boland MP, Staples CR, Thatcher WW, Santos JE. (2017) Effects of supplementation with docosahexaenoic acid on reproduction of dairy cows. Reproduction 153:707–723. [DOI] [PubMed] [Google Scholar]
- Stockton SD, Jr, Gomes I, Liu T, Moraje C, Hipólito L, Jones MR, Ma’ayan A, Morón JA, Li H, Devi LA. (2015) Morphine regulated synaptic networks revealed by integrated proteomics and network analysis. Mol Cell Proteomics 14:2564–2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueta M, Kawai T, Yokoi N, Akira S, Kinoshita S. (2011) Contribution of IPS-1 to polyI:C-induced cytokine production in conjunctival epithelial cells. Biochem Biophys Res Commun 404:419–423. [DOI] [PubMed] [Google Scholar]
- Ujcikova H, Hlouskova M, Cechova K, Stolarova K, Roubalova L, Svoboda P. (2017) Determination of μ-, δ- and κ-opioid receptors in forebrain cortex of rats exposed to morphine for 10 days: comparison with animals after 20 days of morphine withdrawal. PLoS One 12:e0186797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams D, Devi LA. (2010) Escorts take the lead molecular chaperones as therapeutic targets. Prog Mol Biol Transl Sci 91:121–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams JT, Ingram SL, Henderson G, Chavkin C, von Zastrow M, Schulz S, Koch T, Evans CJ, Christie MJ. (2013) Regulation of μ-opioid receptors: desensitization, phosphorylation, internalization, and tolerance. Pharmacol Rev 65:223–254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu J, Faskowitz AJ, Rossi GC, Xu M, Lu Z, Pan YX, Pasternak GW. (2015) Stabilization of morphine tolerance with long-term dosing: association with selective upregulation of mu-opioid receptor splice variant mRNAs. Proc Natl Acad Sci USA 112:279–284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou L, Lovell KM, Frankowski KJ, Slauson SR, Phillips AM, Streicher JM, Stahl E, Schmid CL, Hodder P, Madoux F, et al. (2013) Development of functionally selective, small molecule agonists at kappa opioid receptors. J Biol Chem 288:36703–36716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu ZP, Badisa RB, Palm DE, Goodman CB. (2012) Regulation of rat MOR-1 gene expression after chronic intracerebroventricular administration of morphine. Mol Med Rep 5:513–516. [DOI] [PMC free article] [PubMed] [Google Scholar]