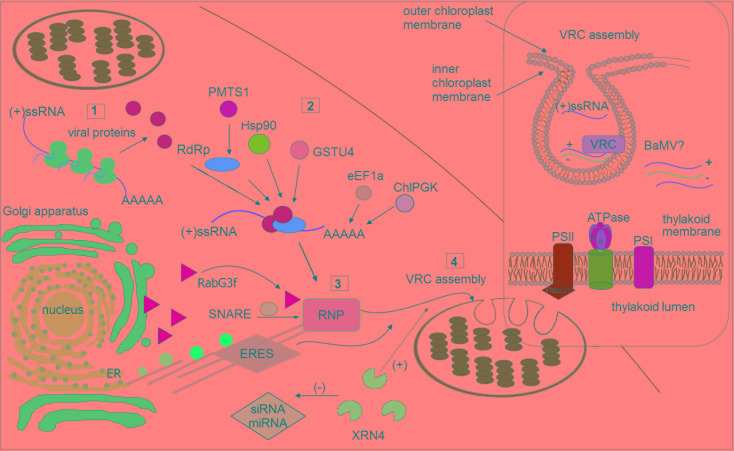
FIGURE 1.
Schematic illustration of selected host factors affecting viral RNA replication in chloroplasts. Synthesis of viral genomic RNA [(+)ssRNA] requires viral proteins: RdRp (pink circle) and auxiliary factors (green circles) that are translated at the early stage of infection (1). Then, virus recruits host factors (2) such as chaperone (Hsp90), methyltransferase (e.g., PMTS1), chloroplast phosphoglycerate kinase (chlPGK), which participate in viral replication complexes (VRC) targeting (in potexvirus). The 3′UTR of viral RNAs (in potexviruses, e.g., BaMV) interacts with several cellular proteins, including elongation factor 1a (eEF1a), glutathione S-transferase (GSTU4), exonuclease (XRN4). Exonuclease negatively regulates the activity of silencing pathway (miRNA/siRNA). Subsequently, the hijacked host factors, viral genomic RNA and viral proteins are trafficked to chloroplast membranes (3). The ribonucleoprotein complex (RNP) is transported from endoplasmic reticulum (ER) exit sites ERES (in potyvirus) or via targeting proteins, such as Rabs or SNARE (in potexvirus) and actomyosin motility system. Chloroplast membranes are later reorganized to form VRC (4). The panel on the right depicts spherule where the viral RNA is synthesized. The host photosynthetic protein PsbO1 from photosystem II is recruited by potyvirus proteins playing a role in its replication (TVBMV, for details refer to the text). In thylakoid membranes, the β subunit of ATPase has been also involved in VRC assembly (potexvirus and tymovirus). Among viruses, viral RNA of BaMV was observed inside the chloroplast. The presented host factors participate in trafficking to the site of replication or affect (directly or indirectly) the synthesis of viral RNA, that is associated with chloroplast.