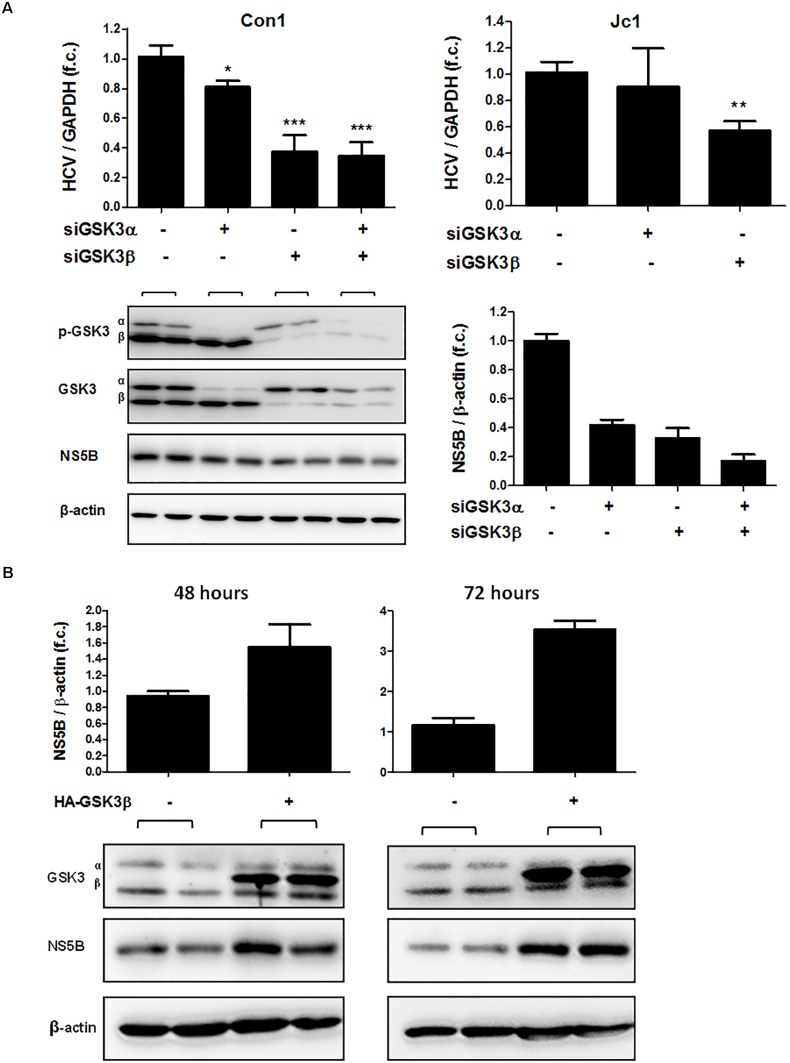
FIGURE 4.
GSK3β is required for HCV replication, not GSK3α. (A) GSK3β silencing suppresses HCV replication. Huh-7.5 cells harboring subgenomic Con1 replicon (left panel) or replicating the full-length Jc1 clone were transfected with siRNAs of GSK3α (siRNA #6236), β (siRNA #6239) or both for 72 h. HCV RNA levels normalized to GAPDH mRNA are expressed relative to untreated cells. Data are presented as mean ± SEM of two experiments performed in triplicate. Asterisks denote statistically significant differences (∗P-value ≤ 0.05; ∗∗P-value ≤ 0.005; ∗∗∗P-value ≤ 0.0005). Immunoblot analysis was carried out for GSK3α/β to confirm gene silencing efficiency, and for HCV NS5B to assess suppression of HCV replication on the protein level (A, lower panel). β-actin was detected as a loading control. NS5B signal intensities were normalized to β-actin for quantification and were presented as mean ± SEM of signal intensities of duplicate bands. (B) GSK3β overexpression promotes HCV expression. Huh-7.5 cells harboring Con1 replicon were transfected with pcDNA3 plasmid expressing HA-tagged wild-type GSK3β (HA-GSK3β) for 48 and 72 h. Immunoblot analysis was carried out for GSK3α/β to confirm transfection efficiency. The overexpression was confirmed by visualizing HA-GSK3β distinct bands with a slightly higher molecular weight than that of the endogenous GSK3β. HCV NS5B protein was detected to assess HCV replication level and β-actin was detected as a loading control. NS5B signal intensities were normalized to β-actin for quantification and were presented as mean ± SEM of signal intensities of duplicate bands.