Abstract
The unique ability of retroviruses to integrate genes into host genomes is of great value for long-term expression in gene therapy, but only when integrations occur at safe genomic sites. To reap the benefit of using retroviruses without severe detrimental effects, we developed several murine leukemia virus (MLV)-based gammaretroviral vectors with safer integration patterns by perturbing the structure of the integrase via insertion of DNA-binding zinc-finger domains (ZFDs) into an internal position of the enzyme. ZFD insertion significantly reduced the inherent, strong MLV integration preference for genomic regions near transcriptional start sites (TSSs), which are the most dangerous spots. The altered retroviral integration pattern was related to increased formation of residual primer-binding site sequences at the 3′ end of proviruses. Several ZFD insertion mutants showed lower frequencies of integrations into the TSS genome regions when having the residual primer-binding site sequences in the proviruses. Our findings not only can extend the use of retroviruses in biomedical applications, but also provide a glimpse into the mechanisms underlying retroviral integration.
Keywords: gammaretroviral vectors, DNA-binding protein domain, integrase, primer-binding site, transcriptional start sites
Introduction
Retroviral vectors, which are mostly based on the murine leukemia virus (MLV), a gammaretrovirus, have been applied in numerous gene therapy clinical trials because of their advantageous characteristics.1, 2, 3, 4 Around 70% of recent gene therapy trials have used viral vectors, approximately 35% and 40% of which employed retroviral and adenoviral vectors, respectively.3 In 2016, the first MLV-based gammaretroviral gene therapy drug, Strimvelis, was approved for the treatment of a rare disease, severe combined immunodeficiency due to adenosine deaminase deficiency.5 Gammaretroviral vectors can carry quite large genetic units of up to 8 kb and cause no significant immune response. Especially, MLV-based gammaretroviral vectors are well studied, and the production of these vectors is simpler than that of lentiviral vectors.4 Most importantly, unlike other vectors, retroviral vectors widely including lentiviral vectors, allow stable gene expression via integration into host chromosomes. However, MLV-based retroviral and HIV-1-based lentiviral vectors, two frequently used vectors, preferentially integrate in genomic regions upstream of genes, mainly transcriptional start sites (TSSs) regions, and within genes, respectively.6 This feature can cause detrimental effects when retroviral vectors integrate upstream of oncogenes and when lentiviral vectors integrate into tumor-suppressor genes. For example, the strong integration preference of MLV-based gammaretroviral vectors for genomic regions near TSSs can lead to uncontrolled growth of transduced human cells through upregulation of downstream oncogenes.7, 8 This oncogenic potential may limit the use of gammaretroviral vectors in clinical applications.
Substantial effort has been made to better understand how retroviruses and lentiviruses select integration sites in mammalian genomes.9, 10, 11, 12, 13, 14, 15, 16, 17, 18, 19 A few cellular proteins with bromodomain and extraterminal domain, and lens epithelium-derived growth factor have been found to tether MLV and HIV-1 to their inherently preferred genomic regions near TSSs and within genes, respectively.10, 11, 12, 13, 14, 16, 18 Based on these findings, several trials, often involving viral integrase engineering, have been conducted to alter the retroviral integration patterns by interfering with the interactions between the cellular tethering factors and the virus, or by conferring viruses the ability to interact with new host proteins.20, 21, 22, 23 In particular, MLV integrations around TSSs have been successfully reduced, but still occur at significantly high frequencies, indicating that there might be other, unknown mechanisms underlying the integration process.20 However, searching the complete MLV integration mechanisms and finding ways to block all the critical mechanisms require substantial time and effort. In this work, instead, we developed a straightforward method that can be generally applied to make safer retroviral and lentiviral vectors. This method involves the perturbation of the structure of integrase, which plays key roles in determining retroviral and lentiviral integration patterns.
Results and Discussion
Retroviral Vectors with Integrase-Zinc-Finger Domain Fusion Proteins
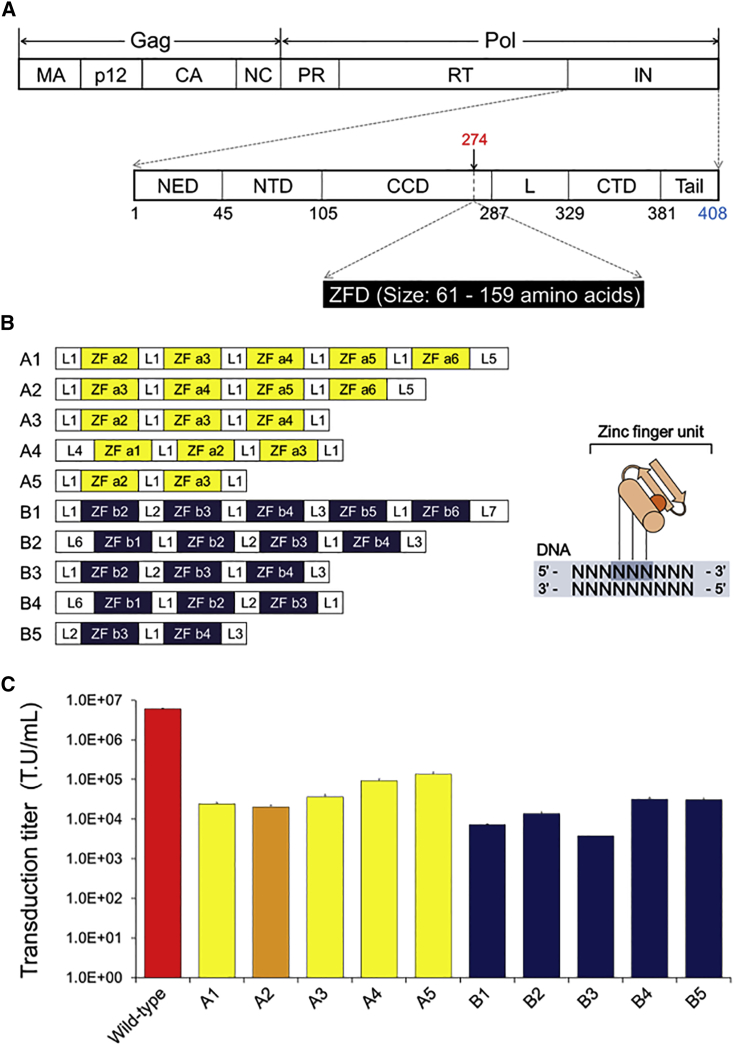
We aimed to develop safer MLV-based gammaretroviral vectors without significant integration preference for TSSs. To this end, we perturbed the structure of integrase by inserting DNA-binding protein domains into an internal site (in front of the 274th amino acid residue; Figure 1A) of the enzyme, which consists of 408 amino acid residues. We expected the consequent changes in the integrase structure to alter the retroviral integration pattern, as generally observed in enzyme structure-function relationships.24, 25 In addition, the DNA-binding property of the inserted domains might support association of the integrase with host genomic DNA.26 As DNA-binding domains, 10 zinc-finger domains (ZFDs) containing between two and five finger units were used (A1–A5 and B1–B5; Figure 1B; Table S1). Because of the short length of the ZFD and the potential alteration of ZFD folding by the flanking viral protein domains, site-specific targeted integration could not be aimed for.
Figure 1.
Constructs of Integrase-ZFD Fusion Proteins and Transduction Titers of Mutant Vectors Harboring the Fusion Proteins
(A) ZFD insertion site within MLV integrase. The top panel depicts MLV Gag-Pol precursor protein, and the bottom panel depicts MLV integrase. Numbers in black denote the starting amino acid residues of each integrase domain. ZFDs were inserted within the integrase at position 274. (B) Constructs of 10 ZFDs that were inserted into MLV integrase. Each ZFD is composed of two to five zinc-finger units. In general, each zinc-finger unit binds to a 3-bp DNA sequence. L1-L7: linker peptides. (C) Transduction titers of MLV-based vectors. HEK293T cells were transduced, and the numbers of EGFP-positive cells were determined by flow cytometry 7 days post-transduction to quantify the transduction titers of viral vectors. Error bars represent the SE of three independent transduction events with an individual vector sample. CCD, catalytic core domain; CTD, C-terminal domain; L, linker; NED, N-terminal extension domain; NTD, N-terminal domain.
Each ZFD was inserted in the MLV vector, resulting in 10 mutant MLV vectors (Figure 1B). Despite the protein insertion, these mutant vectors could still transduce human cells at significant levels, although the transduction titers were inevitably reduced by these mutations, 45.0- to 1,624.8-fold compared with that of the wild-type vector (Figure 1C). A marker protein, EGFP, was expressed at similar levels by wild-type and mutant vectors when the fractions of transduced cells were equivalent (e.g., wild-type versus two mutant vectors [A4 and A5] in Figure S1). Distinct gene expression patterns among the mutant vectors were not recognized in preliminary experiments (J.-E.L., Y.Y., and K. Lim, unpublished data). In contrast, the insertion of foreign proteins into most other sites within the integrase, except for the sites in front of the 38th, 274th, and 378th amino acid residues, completely abrogated the transduction ability of MLV-based vectors.27 Thus, it is not easy to establish infectious mutant virus with protein insertion into an internal site of the integrase. The significant reduction in the functional titer (Figure 1C) may limit the use of mutant vectors for gene delivery in vivo. However, these vectors might be useful for ex-vivo transduction of therapeutic genes into cells, followed by isolation of transduced cells, expansion, and reintroduction of the engineered cells into the patient. Furthermore, the transduction titer of the mutant vectors can be increased by engineering of their genomes and structural proteins. For example, our preliminary independent study revealed that promoter modifications, addition of RNA-stabilizing motifs, and shortening of the genome by omitting the unnecessary parts may increase retroviral transduction titers by several folds.
In this preclinical, basic study, we focused on investigating whether retroviral integration patterns can be altered through perturbation of the integrase structure by using HEK cells, which have been widely used in studies on retroviral integrations,28, 29, 30 rather than using primary cells or stem cells, which would be more relevant for clinical applications.
ZFD Insertion Significantly Reduces Retroviral Integration Preference for the TSS Genomic Regions
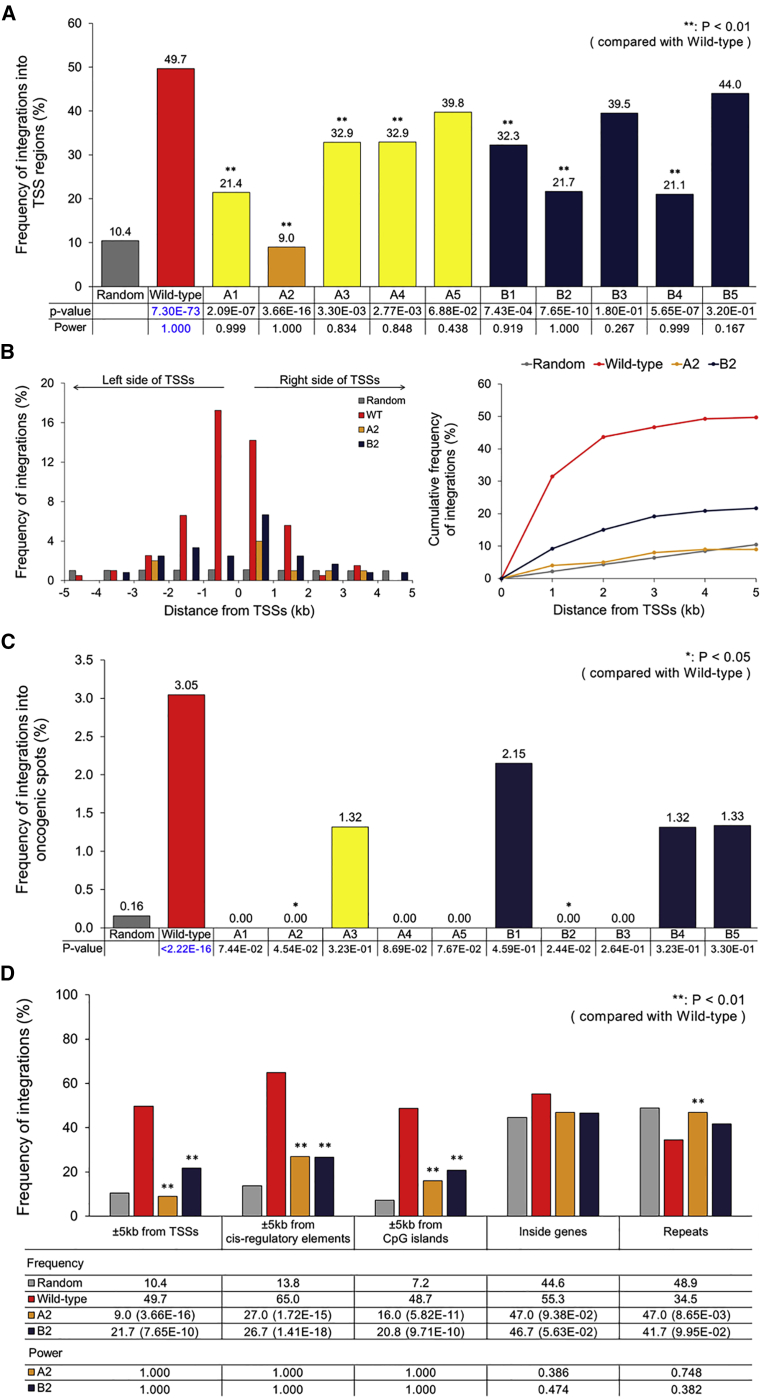
Next-generation sequencing (NGS) and subsequent bioinformatics analysis of host-virus genome junctions was used to assess which human genomic regions harbored retroviral DNAs. Samples were barcoded to allow for multiplex sequencing to reduce the cost while maximizing data yield. We did obtain sufficient non-redundant genome junction reads (Table S2) to detect statistically significantly different integration patterns (p < 0.05 in many cases; Figures 2 and 8). In addition to the determination of the statistical significance for observed differences, post hoc power analysis using the G*Power 3.1 tool31 was conducted to calculate the achieved power given the significance level (α = 0.05), differences, and sample numbers (here, genome junction read numbers). Power values >0.8 (Figures 2 and 8) indicated that we had a sufficient number of genome junction reads to statistically confirm the integration pattern differences for the corresponding cases.31
Figure 2.
Integration Patterns of Mutant Vectors with Integrase-ZFD Proteins in the Human Genome
(A) Frequency of retroviral integrations into the genomic regions within 5 kb of TSSs in the human genome. HEK293T cells were transduced with MLV-based retroviral vectors. Random integrations were additionally computationally generated, and the relevant frequencies of random and retroviral integrations into the TSS regions were quantified, all using the QuickMap tool (Gene Therapy Safety Group50). Statistical significance for the differences between wild-type and mutant integration patterns is indicated by p values (shown in black) that were obtained by the chi-square test. The corresponding statistical power values (see main text) were calculated using G*Power 3.1.31 Statistical significance for the difference between the random and wild-type patterns is indicated by p values (shown in blue) that were obtained by the chi-square test. Corresponding statistical power values were calculated using G*Power 3.1. (B) Left panel: experimentally observed retroviral integrations are spatially distributed within a window of 5 kb upstream and downstream of TSSs in the human genome. Computationally obtained random integrations are also presented over this genomic window. Right panel: cumulative integration frequencies over the indicated distance from TSSs. The frequencies based on total integrations (y coordinate of a point) within x kb from TSSs (x coordinate of a point) are marked in the figure. (C) Frequency of retroviral integrations into genomic sites near TSSs within 5 kb of oncogenes. The frequency was determined by considering the cancer gene census of the Wellcome Trust Sanger Institute.33, 50 For each mutant vector, the hypothesis that it integrates into the dangerous genomic sites at a lower frequency than the wild-type vector was statistically tested by using a binomial test, and the corresponding p value is shown in black. For the wild-type vector, the hypothesis that it integrates into the dangerous sites at a lower frequency than that expected by random chance was tested by using a binomial test, and the corresponding p value is shown in blue. The p value of 2.2E−16 is the smallest p value that can be computed in the statistical package R; p values below this value are all displayed as <2.2E−16 in the package. (D) Frequency of retroviral integrations into different human genomic regions (for wild-type, A2, and B2 mutants). Frequencies of random and experimental retroviral integrations into different genomic regions were quantified, all using the QuickMap tool. Statistical significance of the differences between wild-type and mutant integration patterns is indicated by p values that were obtained by the chi-square test. p values for the comparison with wild-type are shown in parentheses. The corresponding statistical power values were calculated using G*Power 3.1.31 Statistical significance for the difference between the random and wild-type patterns was also quantified using the same approach; p = 7.30E−73 (for TSSs), 1.32E−96 (for cis-regulatory elements), 5.34E−113 (for CpG islands), 2.47E−03 (for Inside genes), and 5.58E−05 (for Repeats).
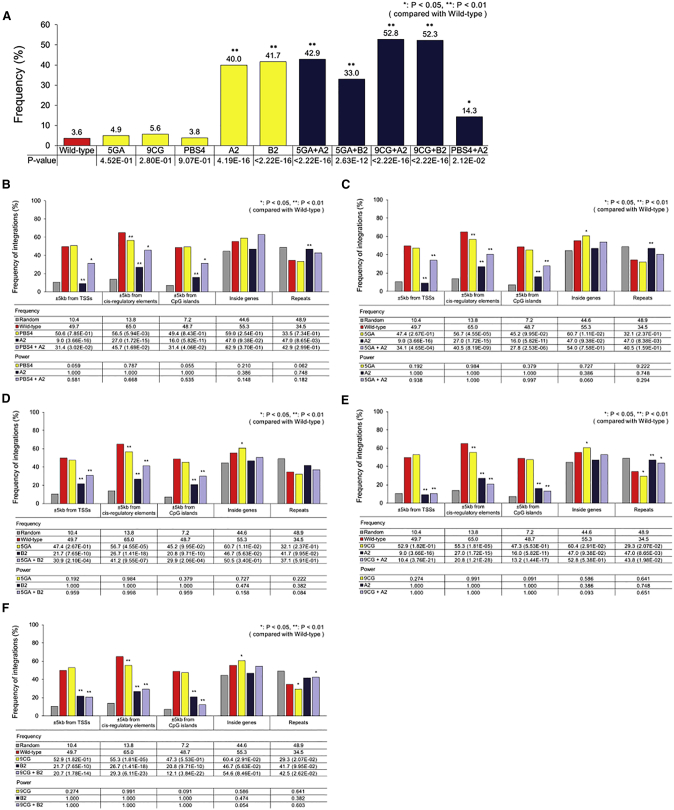
Figure 8.
Integration Patterns of Double Mutants
(A) Frequency of proviruses with residual primer-binding site sequences for single and double mutants. Statistical significance for the difference in residual primer-binding site sequence frequencies between wild-type and mutant vectors is indicated by p values that were obtained by the chi-square test. (B) Frequency of retroviral integrations into different human genomic regions (for PBS4, A2, and PBS4+A2 mutants). Random integrations were computationally generated using the QuickMap tool. The frequencies of random and experimental retroviral integrations into different genomic regions were determined using QuickMap. Statistical significance of the differences between wild-type and mutant integration patterns is indicated by p values that were obtained by the chi-square test. p values for the comparison with wild-type are shown in parentheses. The corresponding statistical power values were determined using G*Power 3.1. (C) For 5GA, A2, and 5GA+A2 mutants. (D) For 5GA, B2, and 5GA+B2 mutants. (E) For 9CG, A2, and 9CG+A2 mutants. (F) For 9CG, B2, and 9CG +B2 mutants. (B–F) Frequency data for wild-type and the single-mutant vectors are from Figures 2D, S8, and S9. PBS, primer-binding site.
In accordance with previous studies,7, 9, 32 the wild-type MLV vector showed a very strong integration preference for genomic regions near TSSs, which are dangerous spots for gene therapy applications, with an integration frequency of 49.7%, which was 4.8-fold higher than that expected by random chance (10.4%; p = 7.30E−73 in comparison with the random case; Figure 2A). In stark contrast, the mutant A2 (Figure 1B) integrated into the TSS regions at only 9.0%, which was 5.5-fold lower than the TSS integration frequency of the wild-type vector and equivalent to that expected by random chance (Figure 2A). This observation indicates that insertion of ZFDs into MLV integrase can completely abolish the retroviral near-TSS integration preference. All other mutants, except A5, B3, and B5, also integrated into near-TSS genomic regions significantly less frequently than the wild-type vector, but more frequently than expected by random chance. This result implied that insertion of DNA-binding domains into integrase does not always yield retroviral vectors with safe integration profiles comparable with that of random integrations. Depending on the size and type of the inserted ZFD, the extent of perturbation of the integrase structure may vary.
Furthermore, within a window of 5 kb upstream and downstream of TSSs, wild-type MLV strongly preferred genomic locations very near the TSSs; it integrated within 1 kb from TSSs at the frequency of 31.4% (Figure 2B, left and right panels). This integration preference specifically for the TSS-proximal regions was 14.2-fold higher than that expected by random chance (2.2%). However, the mutant A2 had a modest preference for these specific genomic regions, with an integration frequency of 4.0%, which was only 1.8-fold higher than that expected by random chance, but 7.9-fold lower than that of the wild-type vector (Figure 2B, left and right panels). The random and mutant A2 integrations showed similar cumulative frequencies over the distance from the TSSs (Figure 2B, right panel). On the other hand, the mutant vector B2 produced a cumulative integration frequency profile between those of the random and wild-type vector integrations, indicating it retained a certain level of integration preference for the TSS-proximal regions (Figure 2B, right panel).
Two Mutant Vectors Show Significantly Lower Preference for TSS Regions Near Oncogenes Than Wild-Type MLV Vector
The oncogenic potential of retroviral vectors is closely correlated with the frequency of vector integrations upstream of oncogenes. QuickMap, which uses the Wellcome Trust Sanger Institute census of human cancer genes,33 was used to search oncogenes within 5 kb of TSSs that neighbored retroviral integrations. Wild-type MLV vectors integrated into these dangerous regions at the frequency of 3.05%, which was significantly higher (19.3-fold) than that expected by random chance (0.16%; p < 2.22E−16; Figure 2C). More specifically, the wild-type vector integrations hit the TSS regions that were near six oncogenes, HOXA9, HOXA11, CCDC6, Myc, PSIP1, and PTPN11 (Table S3). In contrast, the mutants A1, A2, A4, A5, B2, and B3 did not integrate into these oncogenic regions (Figure 2C). Using these integration frequency data, we tested the hypothesis that some mutants integrate less into the oncogenic regions than wild-type vector. A2 and B2 mutant integration patterns satisfied this hypothesis with statistical significance (p = 0.045 and 0.024, respectively; Figure 2C). Integration patterns of A1, A4, and A5 supported this hypothesis with moderate levels of significance (p = 0.074, 0.087, and 0.077, respectively; Figure 2C). These results suggested that insertion of ZFDs into integrase might be a promising approach to reduce the oncogenic risk of MLV-based vectors.
Mutant Vectors Have Significantly Reduced Preference for cis-Regulatory Elements and CpG Islands Compared with Wild-Type MLV Vector
Wild-type MLV vector strongly preferred genomic sites near cis-regulatory elements, as well as CpG islands, which are often enriched in regulatory DNA components, with integration frequencies, respectively, being 4.7- and 6.8-fold higher than that by random chance (Figure 2D). However, insertion of ZFDs into the integrase significantly reduced such integration preference by up to 2.4-fold for cis-regulatory elements and 3.0-fold for CpG islands (for the A2 mutant; Figure 2D). While the wild-type vector also showed slightly higher integration preference for genomic regions within genes than that expected by random chance (p = 2.47E−3), the mutants A2 and B2 hit these regions at frequencies similar to that expected by random integrations (Figure 2D). The integration frequencies of the wild-type vector in genomic regions near TSSs, cis-regulatory elements, CpG islands, and within genes (by 39.3%, 51.2%, 41.5%, and 10.7%, respectively, 142.7% in total) were consistently higher than those expected by random integration, whereas the frequency of integrations into repeats (by 14.4%; Figure 2D) was lower than the random frequency. This large mismatch for 142.7% up and 14.4% down compared with the random case in integration preference for multiple genomic regions indicates that the wild-type vector would integrate into the genomic regions having multiple functions at the same time. Such a strong mismatching trend was not observed for the A2 mutant.
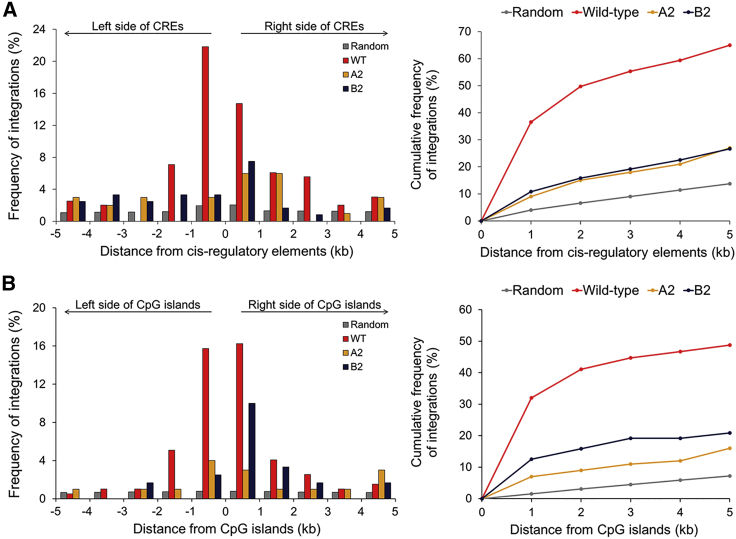
Wild-type MLV vector integrations were also strongly concentrated in genomic sites very near to cis-regulatory elements and CpG islands (within a distance of 1 kb; Figures 3A and 3B). These narrowly distributed wild-type retroviral integrations near regulatory element-rich regions may result in the perturbation of inherent gene expression regulation in host cells. This localized integration centered at regulatory regions was also reduced for the mutant A2 (Figures 3A and 3B).
Figure 3.
Spatial Distribution of Retroviral Integrations around cis-Regulatory Elements and CpG Islands
(A) Left panel: experimentally observed retroviral integrations are spatially distributed within a window 5 kb upstream and downstream of cis-regulatory elements (CREs) that were computationally predicted (referring to cisRED, a genome-scale database51). Right panel: cumulative integration frequencies over the indicated distance from cis-regulatory elements. The frequencies based on total integrations (y coordinate of a point) within x kb from cis-regulatory elements (x coordinate of a point) are marked in the figure. (B) Left panel: experimentally observed retroviral integrations are spatially distributed within a window of 5 kb upstream and downstream of CpG islands. Right panel: cumulative integration frequencies over the indicated distance from CpG islands. The frequencies based on total integrations (y coordinate of a point) within x kb from CpG islands (x coordinate of a point) are marked in the figure. Random integrations were also computationally generated using the QuickMap tool (Gene Therapy Safety Group50). The relevant frequencies of random and experimental retroviral integrations into the genomic regions near the cis-regulatory elements and CpG islands were quantified using the QuickMap tool. The random integrations are also presented over this genomic window.
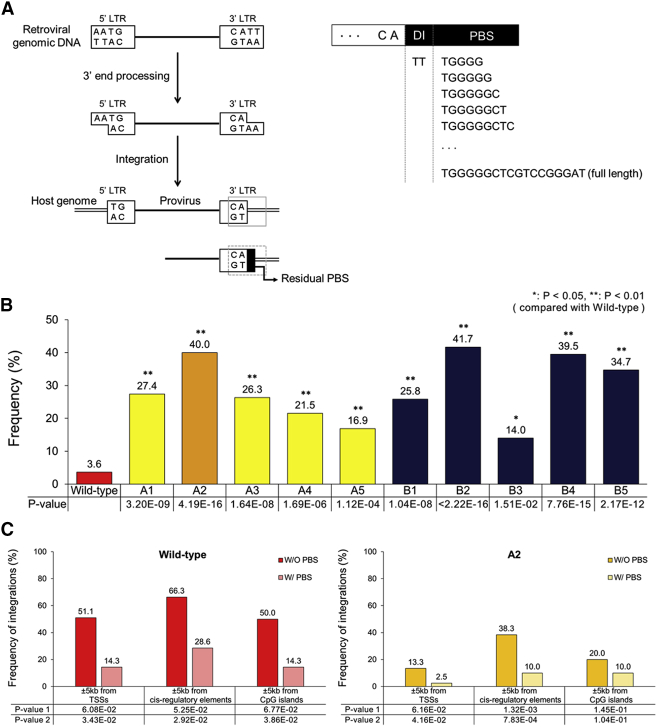
Proviruses of Mutant Vectors Frequently Have Residual Primer-Binding Site Sequences in the 3′ Long Terminal Repeat
Sequence analysis of the host-virus genome junctions revealed that all the mutant vectors frequently produced proviruses with incompletely processed primer-binding site sequences in their 3′ long terminal repeat (LTR) end (Figures 4A and 4B). The formation of primer-binding site sequences at the end of proviruses is thought to be linked to altered activity of the RNase H domain of reverse transcriptase (Figure S2).34, 35 Unexpectedly, proviruses generated from the wild-type vector also had these residual primer-binding site sequences in the 3′ LTR end (Figure 4B). However, the frequency of occurrence of residual primer-binding site sequences per provirus was higher for mutants (14.0%–41.7%) than for the wild-type (3.6%; Figure 4B). Mutants had short residual primer-binding site sequences of variable length (5–18 bp [the entire primer-binding site sequence is 18 bp]; Figure 4A; Figures S3 and S4). Addition of short duplicated DNA sequences from the host genome at the ends of integrated retroviral DNAs often occurs.36 However, it is unclear whether short DNA sequences (<20 bp) flanking the integrated retroviral vector genomes interfere with the expression of transgenes, which are generally located inside the vector genomes. Notably, perturbation of the integrase structure by ZFD insertion led to reduced function of another enzyme, reverse transcriptase. Therefore, our integrase mutants can be considered as class II mutants that can affect reverse transcription, assembly of progeny particles, and other infection steps.37 Similarly, previous studies have reported that mutations of retroviral integrase affected reverse transcription.38, 39, 40
Figure 4.
Genome Integrations of Retroviral Vectors When Having Residual Primer-Binding Site Sequences
(A) Schematic overview of retroviral genomic DNA processing during integration. Left panel: if RNA processing during reverse transcription is incomplete, residual primer-binding site sequences can form in the 3′ LTR end of the provirus (refer to Figure S2). Host genomic DNA is marked as double lines, and viral genomic DNA is denoted as a single, thicker line. Two viral LTRs are denoted with rectangular boxes. The ending sequence of the 3′ LTR is indicated in a gray rectangular box. Residual primer-binding site sequences in the 3′ LTR are indicated by a black filled area. Right panel: examples of residual primer-binding site sequences are listed. In this study, only sequences longer than four bases and starting from the first base of primer-binding site were considered as residual primer-binding site sequences. (B) Frequency of proviruses with residual primer-binding site sequences. Statistical significance for the difference in residual primer-binding site sequence frequencies between wild-type and mutant vectors is indicated by p values that were obtained by the chi-square test. (C) Frequency of retroviral integrations into different human genomic regions in the presence or absence of residual primer-binding site sequences in the provirus 3′ end. Whether the presence of residual primer-binding site sequences is statistically associated with a lower frequency of vector integrations into each type of genomic region was tested using Fisher exact test (p value 1) and Boschloo test (p value 2). The two resulting p values are presented in the frequency plot. Left panel: wild-type vector. Right panel: A2 mutant vector. DI, dinucleotide sequence complementary to the 5′ overhang of dinucleotides that was generated by integrase during 3′ end processing; PBS, primer-binding site.
In addition, although the 5′ overhang of two bases (AA), generated from the 3′ end processing of retroviral genomic DNA (Figure 4A) by integrase,36, 41 was mostly removed for wild-type vector (at the frequency of 94.4%; Table S4), this sequence was less frequently removed for the mutants (at frequencies of 35.5%–67.4%; Table S4). However, the fact that mutant integrases strongly maintained the ability to perform the 3′ end processing of the retroviral genomic DNA corroborated that MLV integrase was functional even with ZFD insertion.
Mutant and Wild-Type Vectors Integrate into the TSS Regions Less Frequently When Having Residual Primer-Binding Site Sequences
To check whether the presence of residual primer-binding site sequences in the provirus 3′ LTR end correlated with the lower tendency of retroviral vectors to integrate into the TSS regions, we divided proviruses into two groups based on the presence or absence of residual primer-binding site sequences and then compared their integration patterns. We first analyzed the mutant vector A2, which showed the lowest integration frequency in the TSS regions (Figure 2A). A2 integrated into genomic regions near TSSs significantly less frequently when having residual primer-binding site sequences than when having no primer-binding site sequence in the 3′ LTR end (2.5% versus 13.3%; Figure 4C, right panel). Similarly, this mutant integrated into cis-regulatory elements significantly less frequently when having residual primer-binding site sequences (Figure 4C, right panel). Integration frequencies for the genomic regions of TSSs, cis-regulatory elements, and CpG islands were partly correlated with the presence of residual primer-binding site sequences also for other mutants (Figures S5 and S6). Interestingly, the wild-type vector showed a similar trend: integrations into the TSS regions, cis-regulatory elements, and CpG islands varied in frequency in accordance with the presence or absence of residual primer-binding site sequences in the provirus 3′ LTR end (Figure 4C, left panel).
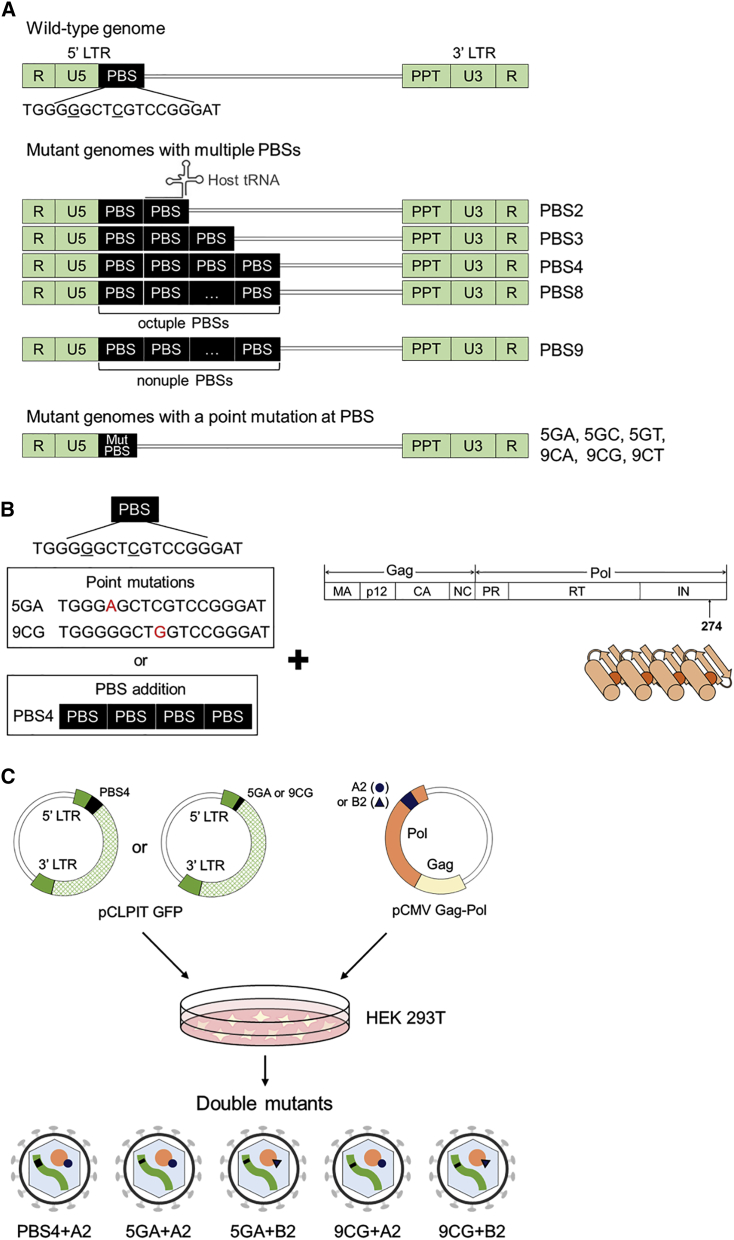
Construction of New Mutants with Modifications of Primer-Binding Site in Their RNA Genome
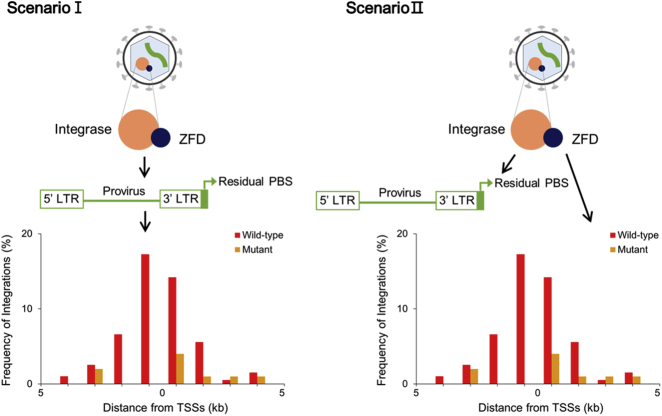
Formation of residual primer-binding site sequences at the 3′ LTR end of reversely transcribed retroviral genomic DNA can be the cause of the shifted integration patterns for the mutants (scenario I; left panel of Figure 5) or another result of the action of an unknown molecular mechanism that altered integration patterns (scenario II; right panel of Figure 5). To assess these two scenarios, we attempted to increase the frequency of residual primer-binding site sequence formation at the 3′ LTR end of retroviral DNA by engineering the MLV RNA genome to have one to eight additional copies of the wild-type primer-binding site sequence in the region flanking the 5′ LTR (Figure 6A). Through host tRNA binding to the newly added primer-binding site(s), the chance of formation of residual primer-binding site sequences at the 3′ LTR end of the retroviral genomic DNA was predicted to increase (refer to Figure S2). To construct additional genome mutants, the fifth and ninth bases of the primer-binding site were also randomly selected to be replaced with one of the three other bases by point mutation (G [5th] to A, C, or T; C [9th] to A, G, or T; Figure 6A). The frequency of occurrence of residual primer-binding site sequences in the provirus 3′ LTR end was slightly increased by 1.7- and 2.1-fold for the PBS2 and 5GT mutants, respectively, compared with that in the wild-type vector (Figure S7; p = 0.185 and 0.0635, respectively). Most mutants with additional primer-binding sites or an altered primer-binding site sequence still showed strong integration preference for genomic regions of TSSs, with frequencies equivalent to that of the wild-type vector (Figures S8 and S9). This result indicates that changes in the primer-binding site sequence alone cannot significantly shift retroviral integrations toward safe genomic regions.
Figure 5.
Two Scenarios to Explain the Experimental Observations in This Study
PBS, primer-binding site.
Figure 6.
Constructs of Mutant Vectors with Modifications of Primer-Binding Site Sequences
(A) Mutations of primer-binding site within the MLV RNA genome. Additional primer-binding sites were added or point mutations were introduced in the primer-binding site. The two bases in the primer-binding site that were point-mutated are underlined. Six mutants with a point mutation were constructed: 5th base of primer-binding site (G) to A (termed 5GA), to C (5GC), to T (5GT), and 9th base of primer-binding site (C) to A (9CA), to G (9CG), and to T (9CT). Host tRNA binding to primer-binding site is also depicted. (B) Schematic diagram of the construction of double mutants. Primer-binding site mutation and ZFD insertion into integrase were applied simultaneously during construction of the double-mutant vectors. (C) Schematic diagram of the production of five double mutants. Mutations of primer-binding site were introduced into pCLPIT GFP, which encodes the vector genome. DNA encoding each zinc-finger domain was introduced into pCMV Gag-Pol-ZFD, which expresses the MLV Gag-Pol polyprotein. PBS, primer-binding site.
In addition, we compared the secondary structures formed by the RNA sequences containing the 5′ functional elements (R, U5, primer-binding site, splice donor site) to evaluate whether incorporation of more primer-binding sites into the viral RNA genome (Figure 6A) could disturb the 5′ stem loops of the genome.42 The secondary RNA structures of the wild-type and primer-binding site mutant viruses were predicted with Mfold.43 The most noticeable change induced by the addition of primer-binding sites was the linear extension of the primer-binding site domain (shown in red in Figure 7). As more primer-binding sites were added, the primer-binding site domain was further extended. In contrast, the secondary structures of other parts were not considerably changed by the addition of primer-binding sites (Figure 7).
Figure 7.
Predicted RNA Secondary Structures
Predicted secondary structures of the 5′ UTR (from R to splice donor site) for wild-type and PBS mutants. The wild-type primer-binding site sequence, newly added primer-binding site sequences, and splice donor site are shown in green, red, and purple, respectively. PBS, primer-binding site.
Mutants with ZFD Insertion Do Not Produce Safe Integration Patterns When Having Additional Primer-Binding Sites in Their RNA Genome
Reasoning that the modification of both the integrase in the viral Gag-Pol polyprotein and primer-binding site in the viral genome may result in safer retroviral integration patterns, we further engineered the MLV vectors. This combined modification approach might additionally aid in assessing the two above-mentioned mechanistic scenarios relevant to the safer integration patterns of the mutants (Figure 5). We inserted one of two ZFDs (A2 or B2; Figure 1B) into the integrase and simultaneously introduced three additional primer-binding sites (PBS4) or a point mutation at the fifth or ninth base of the primer-binding site sequence into viral RNA genome (5GA or 9CG; Figures 6A–6C). With the addition of three primer-binding sites into the genome, proviruses of the double-mutant PBS4+A2 (Figure 6C) had residual primer-binding site sequences in the 3′ LTR end at a significantly lower frequency (14.3%) than those of the mutant A2 (40.0%) without primer-binding site addition (the single mutant, A2) (Figure 8A; p = 0.01). The reduced frequency of residual primer-binding site sequences for this double mutant was associated with a significantly increased frequency of integrations into genomic regions near TSSs (from 9.0% to 31.4%; Figure 8B; the corresponding p value = 3.54E−06). This result indicates that if the primer-binding site domain of the viral RNA genome is not intact, the insertion of ZFD into the integrase may not effectively shift the retroviral integration pattern.
The Presence of Residual Primer-Binding Site Sequences in Provirus Is Not Sufficient for a Safe Retroviral Integration Pattern
With a point mutation at the fifth base of the primer-binding site (G to A; Figures 6A–6C), proviruses of the double mutants 5GA+A2 and 5GA+B2 had residual primer-binding site sequences in the 3′ LTR at frequencies of 42.9% and 33.0%, respectively (Figure 8A), which were equivalent to those of the mutants A2 and B2 without primer-binding site point mutation (40.0% and 41.7%, respectively; Figure 8A). Although they maintained a high frequency of residual primer-binding site sequences, 5GA+A2 and 5GA+B2 integrated into the near-TSS genomic regions at frequencies of 34.1% and 30.9%, respectively (Figures 8C and 8D), which were higher than those of the corresponding single mutants A2 and B2 (9.0% and 21.7%; p = 6.48E−23 and 2.69E−2, respectively) (Figures 8C and 8D). This observation indicates that the occurrence of residual primer-binding site sequences in the 3′ LTR end of proviruses is not sufficient to guarantee safe retroviral integration patterns. In other words, the formation of residual primer-binding site sequences likely results from an unknown molecular mechanism that significantly alters retroviral integration patterns (scenario II in Figure 5), as observed for multiple mutants.
On the other hand, with a point mutation at the ninth base of the primer-binding site (C to G; Figures 6A–6C), proviruses of the double mutants 9CG+A2 and 9CG+B2 had residual primer-binding site sequences in the 3′ LTR at even higher frequencies (52.8% and 52.3%, respectively; Figure 8A) than those of the corresponding single mutants A2 and B2 (40.0% and 41.7%; p = 0.067 and 0.093, respectively; Figure 8A). In contrast with 5GA+A2 and 5GA+B2, 9CG+A2 and 9CG+B2 integrated into near-TSS genomic regions at low frequencies (10.4% and 20.7%, respectively; Figures 8E and 8F), equivalent to those of the corresponding single mutants A2 and B2 (9.0% and 21.7%, respectively; Figures 8E and 8F). Comparison of the integration patterns of the double mutants that had a point mutation at the fifth or ninth position of the primer-binding site further indicates that the ZFD insertion-mediated shift in retroviral integrations toward safer genomic regions requires an intact primer-binding site domain, although the ninth position of the primer-binding site is not a critical position that needs to be conserved.
Conclusions
In this study, we showed that perturbation of the integrase structure by insertion of DNA-binding domains is a simple way to obtain safer retroviral integration patterns. This approach obliterates the need to completely understand the molecular mechanisms that affect retroviral integration patterns and to find effective ways to control these mechanisms. Modification of the integrase significantly reduced the inherent retroviral integration preference for the TSS regions of the human genome to a level expected for random integrations. The concept of integrase structure perturbation can be applied to enhance the safety of lentiviral vectors that have strong integration preference for intergenic regions. Results from preliminary trials indicate that insertion of DNA-binding domains into a few internal positions of the integrase can significantly reduce HIV-1-based vector integrations into genes (Y.Y. and K. Lim, unpublished data). Several decades of efforts in the gene therapy field have resulted in the first approved commercial virus-based gene therapy drugs, Glybera1 and Strimvelis.5 Better control of the safety of retroviral vectors by molecular engineering as shown in this study will allow the production of more effective gene therapy drugs in the near future.
Materials and Methods
Construction of Plasmids Encoding Gag-Pol Mutant Proteins
Two to five zinc-finger units were introduced into an internal site (in front of the 274th amino acid residue) within the MLV integrase as part of the Gag-Pol polyprotein (accession numbers GenBank: J02255, J02256, and J02257) to construct 10 integrase mutant proteins (Figure 1). The DNA molecules encoding the zinc-finger units were amplified via PCR using Phusion High-Fidelity Polymerase (New England Biolabs [NEB], Ipswich, MA, USA). Two plasmids that harbor sequences encoding two zinc-finger complexes, each composed of multiple finger units, were used as PCR templates. The amplified DNA molecules were introduced into the sequence encoding the MLV integrase within the pCMV Gag-Pol plasmid.
Prediction of RNA Secondary Structures
RNA secondary structures of the partial 5′ UTR (from R to splice donor site) were predicted using Mfold43 with the default setting for free-energy minimization. Color coding of the predicted structures was carried out using VARNA.44
Cell Culture
HEK293T cells were cultured in Iscove’s modified Dulbecco’s medium (GIBCO Life Technologies, Carlsbad, CA, USA) supplemented with 10% fetal bovine serum (GIBCO Life Technologies) and 1% penicillin-streptomycin (GIBCO Life Technologies) at 37°C in the presence of 5% CO2.
Vector Packaging
To package MLV-based retroviral vectors, plasmids each encoding the vector genome (pCLPIT GFP, 10 μg), wild-type Gag-Pol polyprotein or mutant Gag-Pol with ZFDs (pCMV Gag-Pol or pCMV Gag-Pol-ZFD, 6 μg), or envelope proteins (pcDNA IVS VSVG, 4 μg) were introduced into HEK293T cells grown in 10-cm dishes via the calcium phosphate-based transfection method. The cell supernatant containing packaged viral particles was harvested twice (1.5 and 2.5 days post-transfection). The harvested supernatant was first filtered through a 0.45-μm syringe filter and then concentrated in a 20% (w/v) sucrose cushion by ultracentrifugation (Optima Ultracentrifuge LE-80K; Beckman Coulter, Brea, CA, USA) using an SW28 rotor at 4°C and 24,000 rpm for 2 hr or an SW41 rotor at 4°C and 25,000 rpm for 1.5 hr. The pellet, containing the virus particles, was resuspended with cooled PBS. Cell supernatant-containing viral particles were also used directly, without concentration.
Titration of Retroviral Transducing Particles
All of the vector genomes (encoded by pCLPIT GFP or modified versions of pCLPIT GFP with mutated primer-binding site sequences) carried the gene encoding EGFP as a reporter gene. Titration of transducing viral particles was performed by transducing HEK293T cells and counting the EGFP-positive cells. Expression of EGFP in cells was measured by flow cytometry on a FACSCanto II flow cytometer (BD Biosciences, San Diego, CA, USA) 7 days post-transduction.
Host-Viral Genome Junction Cloning
HEK293T cells were transduced with wild-type and mutant retroviral vectors at an MOI of less than 1. Transduced cells were expanded for several days and then genomic DNA was isolated from these cells using the QIAamp DNA Mini kit (QIAGEN, Valencia, CA, USA). The genomic DNA was first fragmented with the endonuclease BamHI (NEB) and then linearly amplified by PCR with Taq DNA polymerase (NEB) and a single biotinylated oligonucleotide that binds to the MLV 3′ LTR region (5′-biotin-ATTTGTTAAAGACAGGATATCAGTGGTCCAG-3′). The thermal cycling program was as follows: initial denaturation at 95°C for 5 min, 40 cycles of denaturation at 95°C for 1 min, annealing at 55°C for 45 s, and extension at 72°C for 90 s, final extension at 72°C for 10 min, and cooling at 4°C for 5 min (C1000 thermal cycler; Bio-Rad, Hercules, CA, USA).
After PCR amplification, the product containing the viral sequence was selectively isolated using Dynabeads M-280 Streptavidin (Thermo Fisher Scientific, Carlsbad, CA, USA). To fill the single-stranded region of the isolated DNA product to produce double-stranded (ds)DNA, we conducted an additional DNA synthesis reaction with random hexamer, dNTP, and Klenow enzyme. The dsDNA product was then digested with MseI (NEB), as previously reported,7, 45, 46, 47, 48, 49 and ligated to linker DNA molecules using T4 ligase (NEB). Linker DNA molecules were pre-assembled with two oligos (linker+: 5′-GTAATACGACTCACTATAGGGCTCCGCTTAAGGGAC-3′, linker−: 5′-TAGTCCCTTAAGCGGAG-NH2-3′). The obtained host-viral genome junctions were amplified by PCR with Phusion High-Fidelity Polymerase (NEB) and primers that bind to the viral 3′ LTR or linker (forward, 5′-biotin-GACTTGTGGTCTCGCTGTTCCTTGG-3′, and reverse, 5′- GTAATACGACTCACTATAGGGCTCCGCTTAAG-3′). The thermal cycles were as follows: initial denaturation at 98°C for 2 min, 25 cycles of denaturation at 98°C for 2 min, annealing at 55°C for 90 s, and extension at 72°C for 1 min, final extension at 72°C for 5 min, and cooling at 10°C for 3 min.
NGS of Genome Junctions
The PCR products were analyzed by Illumina NGS. Samples were preprocessed using two consecutive PCRs to add adaptor and index sequences using Phusion High-Fidelity Polymerase (NEB) and the following primers: forward primer for adaptor addition: 5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGGGAGGGTCTCCTCTGAGTGATTGACTACC-3′; reverse primer for adaptor addition: 5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGACTCACTATAGGGCTCCGCTTAAGGGAC-3′; forward primer for index addition: 5′-AATGATACGGCGACCACCGAGATCTACACFFFFFFFFTCGTCGGCAGCGTC-3′; reverse primer for index addition: 5′-CAAGCAGAAGACGGCATACGAGATRRRRRRRRGTCTCGTGGGCTCGG-3′, where “FFFFFFFF” and “RRRRRRRR” are the index sequences.
The thermal cycling condition for adaptor addition was as follows: initial denaturation at 98°C for 2 min, 25 cycles of denaturation at 98°C for 2 min, annealing and extension as a single step at 72°C for 90 s, final extension at 72°C for 5 min, and cooling at 10°C for 3 min. The thermal cycling condition for index addition was as follows: initial denaturation at 98°C for 2 min, 8 cycles of denaturation at 98°C for 12 s, annealing and extension as a single step at 72°C for 90 s, final extension at 72°C for 5 min, and cooling at 10°C for 3 min.
Sequencing using Illumina MiSeq and raw read processing were carried out by Macrogen, a sequencing service provider. To consider only high-quality sequence data, reads with a mean quality score below 20 were filtered out with PRINSEQ-lite (v 0.20.4). Only host-virus genome junction reads containing the viral 3′ LTR sequence (5′-GGAGGGTCTCCTCTGAGTGATTGACTACCCGTCAGCGGGGGTCTTTCA-3′, 48 bp) were captured with total 2-bp mismatch allowance using EMBOSS Needle (v 6.6.0.0) and in-house scripts for downstream analysis. Redundant sequences were also removed during this process. Host sequences from junction reads were mapped to the human genome using the QuickMap tool (Gene Therapy Safety Group50).
Author Contributions
J.-S.N., J.-E.L., K.-H.L., and K.-I.L. designed the experiments; J.-S.N., J.-E.L., and Y.Y. conducted the experiments; all of the authors were involved in analyzing the experimental data; J.-E.L., Y.Y., and K.-I.L. wrote the paper.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This research was supported by the National Research Foundation of Korea (NRF) funded by the Ministry of Science, ICT & Future Planning (grant 2012M3A9B6055200). This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIP) (grant 2011-0030074). This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (grant 2015R1D1A1A01057099).
Footnotes
Supplemental Information includes four tables and nine figures and can be found with this article online at https://doi.org/10.1016/j.omtm.2018.11.001.
Supplemental Information
References
- 1.Naldini L. Gene therapy returns to centre stage. Nature. 2015;526:351–360. doi: 10.1038/nature15818. [DOI] [PubMed] [Google Scholar]
- 2.Schaffer D.V., Koerber J.T., Lim K.I. Molecular engineering of viral gene delivery vehicles. Annu. Rev. Biomed. Eng. 2008;10:169–194. doi: 10.1146/annurev.bioeng.10.061807.160514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Xu X., Tailor C.S., Grunebaum E. Gene therapy for primary immune deficiencies: a Canadian perspective. Allergy Asthma Clin. Immunol. 2017;13:14. doi: 10.1186/s13223-017-0184-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vargas J.E., Chicaybam L., Stein R.T., Tanuri A., Delgado-Cañedo A., Bonamino M.H. Retroviral vectors and transposons for stable gene therapy: advances, current challenges and perspectives. J. Transl. Med. 2016;14:288. doi: 10.1186/s12967-016-1047-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aiuti A., Roncarolo M.G., Naldini L. Gene therapy for ADA-SCID, the first marketing approval of an ex vivo gene therapy in Europe: paving the road for the next generation of advanced therapy medicinal products. EMBO Mol. Med. 2017;9:737–740. doi: 10.15252/emmm.201707573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cattoglio C., Pellin D., Rizzi E., Maruggi G., Corti G., Miselli F., Sartori D., Guffanti A., Di Serio C., Ambrosi A. High-definition mapping of retroviral integration sites identifies active regulatory elements in human multipotent hematopoietic progenitors. Blood. 2010;116:5507–5517. doi: 10.1182/blood-2010-05-283523. [DOI] [PubMed] [Google Scholar]
- 7.Wu X., Li Y., Crise B., Burgess S.M. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]
- 8.Hacein-Bey-Abina S., Von Kalle C., Schmidt M., McCormack M.P., Wulffraat N., Leboulch P., Lim A., Osborne C.S., Pawliuk R., Morillon E. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 9.Lewinski M.K., Yamashita M., Emerman M., Ciuffi A., Marshall H., Crawford G., Collins F., Shinn P., Leipzig J., Hannenhalli S. Retroviral DNA integration: viral and cellular determinants of target-site selection. PLoS Pathog. 2006;2:e60. doi: 10.1371/journal.ppat.0020060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Rijck J., de Kogel C., Demeulemeester J., Vets S., El Ashkar S., Malani N., Bushman F.D., Landuyt B., Husson S.J., Busschots K. The BET family of proteins targets moloney murine leukemia virus integration near transcription start sites. Cell Rep. 2013;5:886–894. doi: 10.1016/j.celrep.2013.09.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sharma A., Larue R.C., Plumb M.R., Malani N., Male F., Slaughter A., Kessl J.J., Shkriabai N., Coward E., Aiyer S.S. BET proteins promote efficient murine leukemia virus integration at transcription start sites. Proc. Natl. Acad. Sci. USA. 2013;110:12036–12041. doi: 10.1073/pnas.1307157110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ciuffi A., Llano M., Poeschla E., Hoffmann C., Leipzig J., Shinn P., Ecker J.R., Bushman F. A role for LEDGF/p75 in targeting HIV DNA integration. Nat. Med. 2005;11:1287–1289. doi: 10.1038/nm1329. [DOI] [PubMed] [Google Scholar]
- 13.Shun M.-C., Raghavendra N.K., Vandegraaff N., Daigle J.E., Hughes S., Kellam P., Cherepanov P., Engelman A. LEDGF/p75 functions downstream from preintegration complex formation to effect gene-specific HIV-1 integration. Genes Dev. 2007;21:1767–1778. doi: 10.1101/gad.1565107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Singh P.K., Plumb M.R., Ferris A.L., Iben J.R., Wu X., Fadel H.J., Luke B.T., Esnault C., Poeschla E.M., Hughes S.H. LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1 integration to highly spliced genes. Genes Dev. 2015;29:2287–2297. doi: 10.1101/gad.267609.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Babaei S., Akhtar W., de Jong J., Reinders M., de Ridder J. 3D hotspots of recurrent retroviral insertions reveal long-range interactions with cancer genes. Nat. Commun. 2015;6:6381. doi: 10.1038/ncomms7381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaFave M.C., Varshney G.K., Gildea D.E., Wolfsberg T.G., Baxevanis A.D., Burgess S.M. MLV integration site selection is driven by strong enhancers and active promoters. Nucleic Acids Res. 2014;42:4257–4269. doi: 10.1093/nar/gkt1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Melamed A., Laydon D.J., Gillet N.A., Tanaka Y., Taylor G.P., Bangham C.R. Genome-wide determinants of proviral targeting, clonal abundance and expression in natural HTLV-1 infection. PLoS Pathog. 2013;9:e1003271. doi: 10.1371/journal.ppat.1003271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Crowe B.L., Larue R.C., Yuan C., Hess S., Kvaratskhelia M., Foster M.P. Structure of the Brd4 ET domain bound to a C-terminal motif from γ-retroviral integrases reveals a conserved mechanism of interaction. Proc. Natl. Acad. Sci. USA. 2016;113:2086–2091. doi: 10.1073/pnas.1516813113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marini B., Kertesz-Farkas A., Ali H., Lucic B., Lisek K., Manganaro L., Pongor S., Luzzati R., Recchia A., Mavilio F. Nuclear architecture dictates HIV-1 integration site selection. Nature. 2015;521:227–231. doi: 10.1038/nature14226. [DOI] [PubMed] [Google Scholar]
- 20.Aiyer S., Swapna G.V., Malani N., Aramini J.M., Schneider W.M., Plumb M.R., Ghanem M., Larue R.C., Sharma A., Studamire B. Altering murine leukemia virus integration through disruption of the integrase and BET protein family interaction. Nucleic Acids Res. 2014;42:5917–5928. doi: 10.1093/nar/gku175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.El Ashkar S., De Rijck J., Demeulemeester J., Vets S., Madlala P., Cermakova K., Debyser Z., Gijsbers R. BET-independent MLV-based vectors target away from promoters and regulatory elements. Mol. Ther. Nucleic Acids. 2014;3:e179. doi: 10.1038/mtna.2014.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hocum J.D., Linde I., Rae D.T., Collins C.P., Matern L.K., Trobridge G.D. Retargeted foamy virus vectors integrate less frequently near proto-oncogenes. Sci. Rep. 2016;6:36610. doi: 10.1038/srep36610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.El Ashkar S., Van Looveren D., Schenk F., Vranckx L.S., Demeulemeester J., De Rijck J., Debyser Z., Modlich U., Gijsbers R. Engineering next-generation BET-independent MLV vectors for safer gene therapy. Mol. Ther. Nucleic Acids. 2017;7:231–245. doi: 10.1016/j.omtn.2017.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Girard E., Marchal S., Perez J., Finet S., Kahn R., Fourme R., Marassio G., Dhaussy A.C., Prangé T., Giffard M. Structure-function perturbation and dissociation of tetrameric urate oxidase by high hydrostatic pressure. Biophys. J. 2010;98:2365–2373. doi: 10.1016/j.bpj.2010.01.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo Q., He Y., Lu H.P. Interrogating the activities of conformational deformed enzyme by single-molecule fluorescence-magnetic tweezers microscopy. Proc. Natl. Acad. Sci. USA. 2015;112:13904–13909. doi: 10.1073/pnas.1506405112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee S., Oh Y., Lee J., Choe S., Lim S., Lee H.S., Jo K., Schwartz D.C. DNA binding fluorescent proteins for the direct visualization of large DNA molecules. Nucleic Acids Res. 2016;44:e6. doi: 10.1093/nar/gkv834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lim K.I., Klimczak R., Yu J.H., Schaffer D.V. Specific insertions of zinc finger domains into Gag-Pol yield engineered retroviral vectors with selective integration properties. Proc. Natl. Acad. Sci. USA. 2010;107:12475–12480. doi: 10.1073/pnas.1001402107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Santoni F.A., Hartley O., Luban J. Deciphering the code for retroviral integration target site selection. PLoS Comput. Biol. 2010;6:e1001008. doi: 10.1371/journal.pcbi.1001008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Moalic Y., Félix H., Takeuchi Y., Jestin A., Blanchard Y. Genome areas with high gene density and CpG island neighborhood strongly attract porcine endogenous retrovirus for integration and favor the formation of hot spots. J. Virol. 2009;83:1920–1929. doi: 10.1128/JVI.00856-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kvaratskhelia M., Sharma A., Larue R.C., Serrao E., Engelman A. Molecular mechanisms of retroviral integration site selection. Nucleic Acids Res. 2014;42:10209–10225. doi: 10.1093/nar/gku769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Faul F., Erdfelder E., Buchner A., Lang A.G. Statistical power analyses using G*Power 3.1: tests for correlation and regression analyses. Behav. Res. Methods. 2009;41:1149–1160. doi: 10.3758/BRM.41.4.1149. [DOI] [PubMed] [Google Scholar]
- 32.Beard B.C., Dickerson D., Beebe K., Gooch C., Fletcher J., Okbinoglu T., Miller D.G., Jacobs M.A., Kaul R., Kiem H.P., Trobridge G.D. Comparison of HIV-derived lentiviral and MLV-based gammaretroviral vector integration sites in primate repopulating cells. Mol. Ther. 2007;15:1356–1365. doi: 10.1038/sj.mt.6300159. [DOI] [PubMed] [Google Scholar]
- 33.Futreal P.A., Coin L., Marshall M., Down T., Hubbard T., Wooster R., Rahman N., Stratton M.R. A census of human cancer genes. Nat. Rev. Cancer. 2004;4:177–183. doi: 10.1038/nrc1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Julias J.G., McWilliams M.J., Sarafianos S.G., Arnold E., Hughes S.H. Mutations in the RNase H domain of HIV-1 reverse transcriptase affect the initiation of DNA synthesis and the specificity of RNase H cleavage in vivo. Proc. Natl. Acad. Sci. USA. 2002;99:9515–9520. doi: 10.1073/pnas.142123199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Julias J.G., McWilliams M.J., Sarafianos S.G., Alvord W.G., Arnold E., Hughes S.H. Mutation of amino acids in the connection domain of human immunodeficiency virus type 1 reverse transcriptase that contact the template-primer affects RNase H activity. J. Virol. 2003;77:8548–8554. doi: 10.1128/JVI.77.15.8548-8554.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim S., Rusmevichientong A., Dong B., Remenyi R., Silverman R.H., Chow S.A. Fidelity of target site duplication and sequence preference during integration of xenotropic murine leukemia virus-related virus. PLoS ONE. 2010;5:e10255. doi: 10.1371/journal.pone.0010255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lu R., Limón A., Devroe E., Silver P.A., Cherepanov P., Engelman A. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 2004;78:12735–12746. doi: 10.1128/JVI.78.23.12735-12746.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wilkinson T.A., Januszyk K., Phillips M.L., Tekeste S.S., Zhang M., Miller J.T., Le Grice S.F., Clubb R.T., Chow S.A. Identifying and characterizing a functional HIV-1 reverse transcriptase-binding site on integrase. J. Biol. Chem. 2009;284:7931–7939. doi: 10.1074/jbc.M806241200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dobard C.W., Briones M.S., Chow S.A. Molecular mechanisms by which human immunodeficiency virus type 1 integrase stimulates the early steps of reverse transcription. J. Virol. 2007;81:10037–10046. doi: 10.1128/JVI.00519-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chakraborty A., Sun G.Q., Mustavich L., Huang S.H., Li B.L. Biochemical interactions between HIV-1 integrase and reverse transcriptase. FEBS Lett. 2013;587:425–429. doi: 10.1016/j.febslet.2012.12.007. [DOI] [PubMed] [Google Scholar]
- 41.Scottoline B.P., Chow S., Ellison V., Brown P.O. Disruption of the terminal base pairs of retroviral DNA during integration. Genes Dev. 1997;11:371–382. doi: 10.1101/gad.11.3.371. [DOI] [PubMed] [Google Scholar]
- 42.Mougel M., Tounekti N., Darlix J.-L., Paoletti J., Ehresmann B., Ehresmann C. Conformational analysis of the 5′ leader and the gag initiation site of Mo-MuLV RNA and allosteric transitions induced by dimerization. Nucleic Acids Res. 1993;21:4677–4684. doi: 10.1093/nar/21.20.4677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Darty K., Denise A., Ponty Y. VARNA: interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hashemi F.B., Barreto K., Bernhard W., Hashemi P., Lomness A., Sadowski I. HIV provirus stably reproduces parental latent and induced transcription phenotypes regardless of the chromosomal integration site. J. Virol. 2016;90:5302–5314. doi: 10.1128/JVI.02842-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Demeulemeester J., Vets S., Schrijvers R., Madlala P., De Maeyer M., De Rijck J., Ndung’u T., Debyser Z., Gijsbers R. HIV-1 integrase variants retarget viral integration and are associated with disease progression in a chronic infection cohort. Cell Host Microbe. 2014;16:651–662. doi: 10.1016/j.chom.2014.09.016. [DOI] [PubMed] [Google Scholar]
- 47.Derse D., Crise B., Li Y., Princler G., Lum N., Stewart C., McGrath C.F., Hughes S.H., Munroe D.J., Wu X. Human T-cell leukemia virus type 1 integration target sites in the human genome: comparison with those of other retroviruses. J. Virol. 2007;81:6731–6741. doi: 10.1128/JVI.02752-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vranckx L.S., Demeulemeester J., Debyser Z., Gijsbers R. Towards a safer, more randomized lentiviral vector integration profile exploring artificial LEDGF chimeras. PLoS ONE. 2016;11:e0164167. doi: 10.1371/journal.pone.0164167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Moiani A., Paleari Y., Sartori D., Mezzadra R., Miccio A., Cattoglio C., Cocchiarella F., Lidonnici M.R., Ferrari G., Mavilio F. Lentiviral vector integration in the human genome induces alternative splicing and generates aberrant transcripts. J. Clin. Invest. 2012;122:1653–1666. doi: 10.1172/JCI61852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Appelt J.U., Giordano F.A., Ecker M., Roeder I., Grund N., Hotz-Wagenblatt A., Opelz G., Zeller W.J., Allgayer H., Fruehauf S., Laufs S. QuickMap: a public tool for large-scale gene therapy vector insertion site mapping and analysis. Gene Ther. 2009;16:885–893. doi: 10.1038/gt.2009.37. [DOI] [PubMed] [Google Scholar]
- 51.Robertson G., Bilenky M., Lin K., He A., Yuen W., Dagpinar M., Varhol R., Teague K., Griffith O.L., Zhang X. cisRED: a database system for genome-scale computational discovery of regulatory elements. Nucleic Acids Res. 2006;34:D68–D73. doi: 10.1093/nar/gkj075. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.