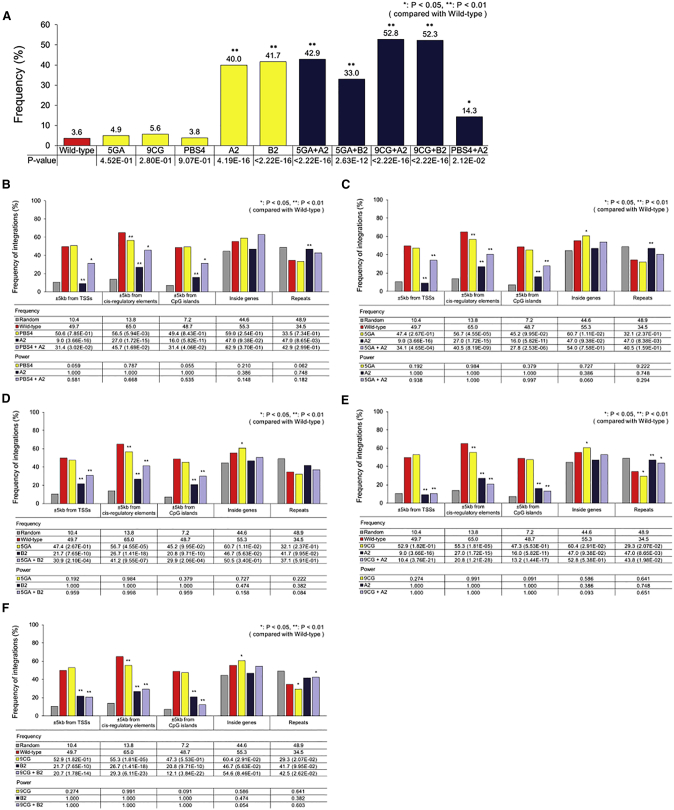
Figure 8.
Integration Patterns of Double Mutants
(A) Frequency of proviruses with residual primer-binding site sequences for single and double mutants. Statistical significance for the difference in residual primer-binding site sequence frequencies between wild-type and mutant vectors is indicated by p values that were obtained by the chi-square test. (B) Frequency of retroviral integrations into different human genomic regions (for PBS4, A2, and PBS4+A2 mutants). Random integrations were computationally generated using the QuickMap tool. The frequencies of random and experimental retroviral integrations into different genomic regions were determined using QuickMap. Statistical significance of the differences between wild-type and mutant integration patterns is indicated by p values that were obtained by the chi-square test. p values for the comparison with wild-type are shown in parentheses. The corresponding statistical power values were determined using G*Power 3.1. (C) For 5GA, A2, and 5GA+A2 mutants. (D) For 5GA, B2, and 5GA+B2 mutants. (E) For 9CG, A2, and 9CG+A2 mutants. (F) For 9CG, B2, and 9CG +B2 mutants. (B–F) Frequency data for wild-type and the single-mutant vectors are from Figures 2D, S8, and S9. PBS, primer-binding site.